---
title: Pair Plots
date: last-modified
lightbox: true
toc: true
page-layout: full
format:
html:
grid:
sidebar-width: 200px
body-width: 1000px
margin-width: 200px
gutter-width: 1.0rem
toc-depth: 3
code-fold: true
code-tools: true
execute:
warning: false
eval: true
---
```{r}
pacman::p_load(dplyr,purrr,tidyr,ggplot2, here, patchwork,
conflicted,stringr, gt, knitr, kableExtra,
lubridate,ggh4x,jsonlite,httr, openai)
walk(c("dplyr"), conflict_prefer_all, quiet = TRUE)
options(digits=2, scipen=999, dplyr.summarise.inform=FALSE)
#walk(c("fun_plot"), ~ source(here::here(paste0("R/", .x, ".R"))))
mc24_proto <- read.csv(here("Stimulii","mc24_prototypes.csv")) |> mutate(set=paste0(sbjCode,"_",condit))
sbj_cat <- read.csv(here("data","mc24_sbj_cat.csv"))
dfiles <- list(path=list.files(here::here("data/dotSim_data"),full.names=TRUE))
d <- map_dfr(dfiles$path, ~read.csv(.x))
d <- map_dfr(dfiles$path, ~{read.csv(.x) |>
mutate(sfile=tools::file_path_sans_ext(basename(.x)))}) |>
select(-trial_index, -internal_node_id,-trial_type) |>
mutate(set = paste(str_extract(item_label_1, "^\\d+"),
str_extract(item_label_1, "[a-z]+"), sep = "_")) |>
mutate(pair_label = paste0(item_label_1,"_",item_label_2)) |>
relocate(sbjCode,date,set,pair_label,trial,item_label_1,item_label_2,response,rt)
setCounts <- d |>
pivot_longer(cols=c(item_label_1, item_label_2), names_to="item_label", values_to="item") |>
group_by(set) |> summarise(n=n_distinct(sbjCode),resp=mean(response),sd=sd(response)) |> arrange(desc(n))
# length(unique(mc_proto$set)) # 304
setCounts2 <- mc24_proto |> group_by(set) |>
slice_head(n=1) |>
select(id,file,set) |>
left_join(setCounts,by="set") |>
mutate(n = ifelse(is.na(n), 0, n), .groups="drop") |>
arrange(n) |> ungroup()
pairCounts <- d |>
group_by(pair_label,set) |>
summarise(n=n(),mean_resp=mean(response),sd=sd(response)) |>
arrange(mean_resp) |> # arrange(desc(n)) |>
ungroup()
# saveRDS(pairCounts,here::here("data","pairCounts.rds"))
#pairCounts <- readRDS(here("data/pairCounts.rds")) |> arrange(mean_resp)
```
```{r}
#| fig-width: 12
#| fig-height: 9
pair_plot <- function(Pair,item_labels=FALSE){
df <- d |> filter(pair_label==Pair) |> slice_head(n=1)
dim1=30
x_limits <- c(-dim1, dim1) # Set fixed x-axis limits
y_limits <- c(-dim1, dim1) # Set fixed y-axis limits
pat1 <- df %>%
mutate(pattern_1 = purrr::map(pattern_1, jsonlite::fromJSON)) %>%
unnest(pattern_1) %>%
mutate(y=-y, pat=item_label_1) |> select(pair_label,x,y,pat)
pat2 <- df %>%
mutate(pattern_2 = purrr::map(pattern_2, jsonlite::fromJSON)) %>%
unnest(pattern_2) %>%
mutate(y=-y, pat=item_label_2) |> select(pair_label,x,y,pat)
pat <- rbind(pat1,pat2)
pat |>
ggplot(aes(x = x, y = y #,fill=pat,col=pat
)) +
geom_point(alpha=2,size=.8) +
coord_cartesian(xlim = x_limits, ylim =y_limits) +
theme_minimal() +
facet_wrap(~pat,ncol=2,scales = "fixed") + #axes="all"
#theme_blank +
theme_void() +
theme(strip.text = element_text(size = 7,hjust=.5),
#panel.spacing.x=unit(-8.5, "lines"),
#strip.background = element_rect(colour = "black", linewidth = 2),
strip.text.x = if(item_labels) element_text() else element_blank(), # remove pattern labels
panel.border = element_blank(), # Remove borders around facets
legend.position = "none",
plot.background = element_rect(fill = "white"),
panel.background = element_rect(fill = "white"),
#axis.line.y = element_line(colour = "black", linewidth = .1)
) +
xlim(x_limits[1], x_limits[2]) + # Set x-axis limits explicitly
ylim(y_limits[1], y_limits[2]) # Set y-axis limits explicitly
}
save_pair <- function(Pair) {
p <- pair_plot(Pair)
ggsave(filename = paste0(here("Stimulii/pair_images/"),Pair, ".png"), plot = p,width = 4, height = 3, dpi = 200)
}
plot_hist_pair <- function(Pair) {
d |> filter(pair_label==Pair) |>
ggplot(aes(x = response)) +
geom_histogram(binwidth=1,fill = 'dodgerblue4') +
scale_x_continuous(breaks=seq(1, 9, by = 1)) +
coord_cartesian(xlim = c(1, 9)) +
theme_minimal() +
# theme_void() +
theme(#axis.title.x=element_blank(),
#axis.title.y=element_blank(),
plot.title=element_text(face="plain",size=14,hjust=.5,vjust=1),
axis.text.x=element_text(size=12)) +
labs(x="Similarity Rating (low to high)", y= "Response Frequency",title="Response Distribution")
}
pair_hist_plot <- function(Pair,item_labels=FALSE,pair_title=""){
df <- d |> filter(pair_label==Pair) |> slice_head(n=1)
pat1 <- df %>%
mutate(pattern_1 = purrr::map(pattern_1, jsonlite::fromJSON)) %>%
unnest(pattern_1) %>%
mutate(y=-y, pat=item_label_1) |> select(pair_label,x,y,pat)
pat2 <- df %>%
mutate(pattern_2 = purrr::map(pattern_2, jsonlite::fromJSON)) %>%
unnest(pattern_2) %>%
mutate(y=-y, pat=item_label_2) |> select(pair_label,x,y,pat)
hist <- plot_hist_pair(Pair)
pat <- rbind(pat1,pat2)
dim1=30
max_dim <- max(max(abs(pat$x)),max(abs(pat$y)))+5
x_limits <- c(-max_dim, max_dim) # Set fixed x-axis limits
y_limits <- c(-max_dim, max_dim) # Set fixed y-axis limits
pat_p <- pat |>
ggplot(aes(x = x, y = y #,fill=pat,col=pat
)) +
geom_point(alpha=2,size=1.5) +
coord_cartesian(xlim = x_limits, ylim =y_limits) +
theme_minimal() +
facet_wrap(~pat,ncol=2,scales = "fixed") + #axes="all"
#theme_blank +
theme(#strip.text = element_text(size = 7,hjust=.5),
#panel.spacing.x=unit(-8.5, "lines"),
#strip.background = element_rect(colour = "black", linewidth = 2),
strip.text.x = if(item_labels) element_text() else element_blank(), # remove pattern labels
panel.border = element_blank(), # Remove borders around facets
legend.position = "none",
plot.background = element_rect(fill = "white"),
#panel.background = element_rect(fill = "white"),
#axis.line.y = element_line(colour = "black", linewidth = .1)
) +
labs(title=pair_title) +
theme_void() +
theme(strip.text.x = if(item_labels) element_text() else element_blank(),
plot.title=element_text(face="plain",size=14,hjust=.5,vjust=1) )+
xlim(x_limits[1], x_limits[2]) + # Set x-axis limits explicitly
ylim(y_limits[1], y_limits[2]) #y_limits[2]) # Set y-axis limits explicitly
pat_p + hist + plot_layout(widths = c(2.0, 1))
}
```
```{r}
#| fig-width: 11
#| fig-height: 9
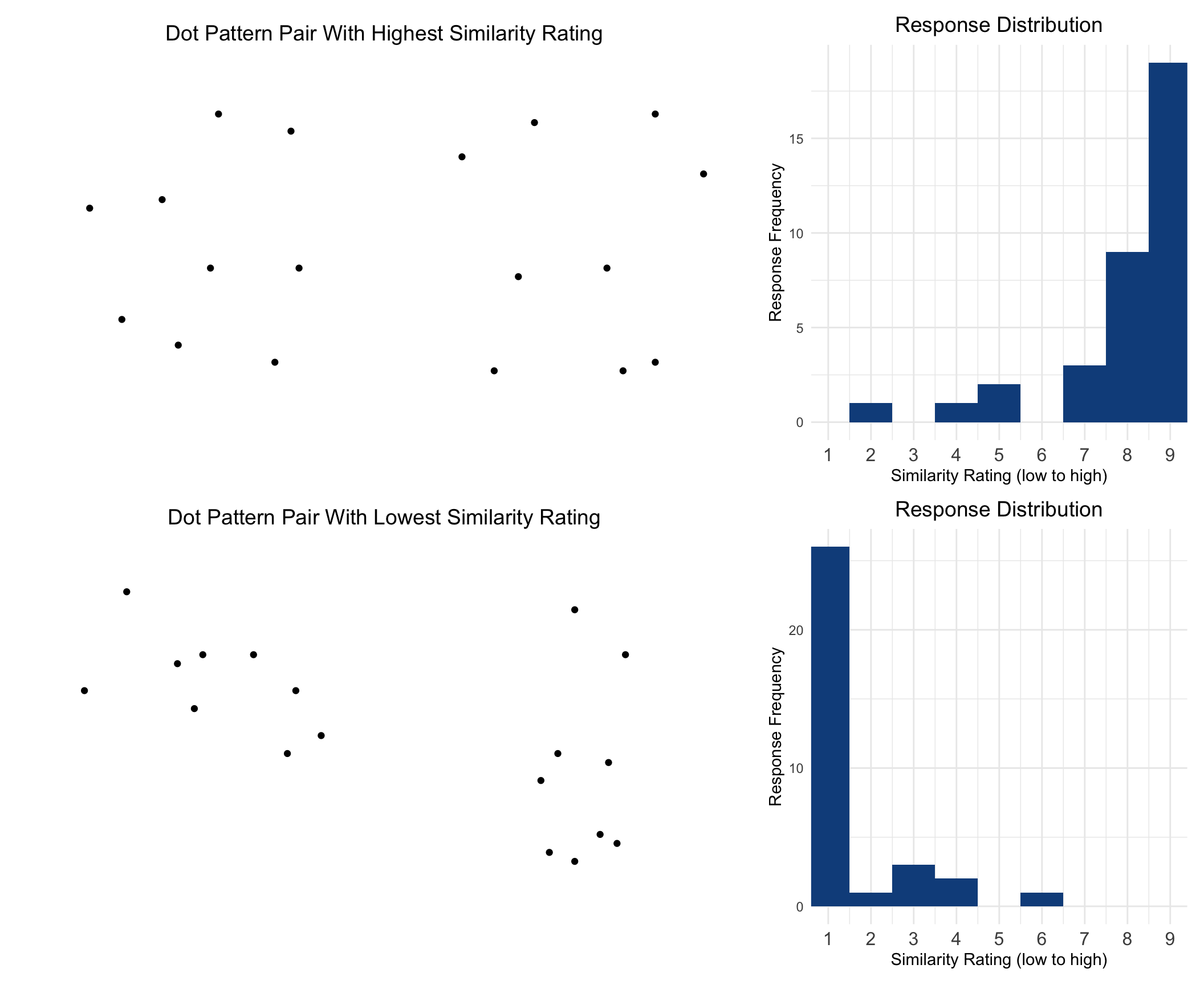
ph <- pairCounts |>
slice_tail(n = 1) |>
arrange(desc(mean_resp)) |>
pull(pair_label) |>
map(~pair_hist_plot(.x, item_labels = FALSE,pair_title="Dot Pattern Pair With Highest Similarity Rating")) |>
wrap_plots(ncol = 1)
pl <- pairCounts |>
slice_head(n = 1) |>
pull(pair_label) |>
map(~pair_hist_plot(.x, item_labels = FALSE,pair_title="Dot Pattern Pair With Lowest Similarity Rating")) |>
wrap_plots(ncol = 1)
pair_extremes <- ph / pl
# ggsave(filename=paste0(here::here(),"/assets/dot_extremes.png"),plot=pair_extremes,bg='transparent',dpi=400)
pair_extremes
#plot_annotation(title = "Dot Pattern Pair With Highest Similarity Rating")
# plot 3 pairs from the middle of pairCounts
# pairCounts |>
# slice(ceiling(n()/2)-1:3) |>
# pull(pair_label) |>
# map(~pair_plot(.x, item_labels = TRUE)) |>
# wrap_plots(ncol = 1) +
# plot_annotation(title = "Middle Rated Pairs")
#
# combined_df |>
# pull(pair_label) |>
# walk(~ {
# p <- pair_plot(.x, item_labels = TRUE)
# ggsave(filename = paste0(here("llm/ex_images/"), .x, ".png"), plot = p, width = 4, height = 3, dpi = 200)
# })
```
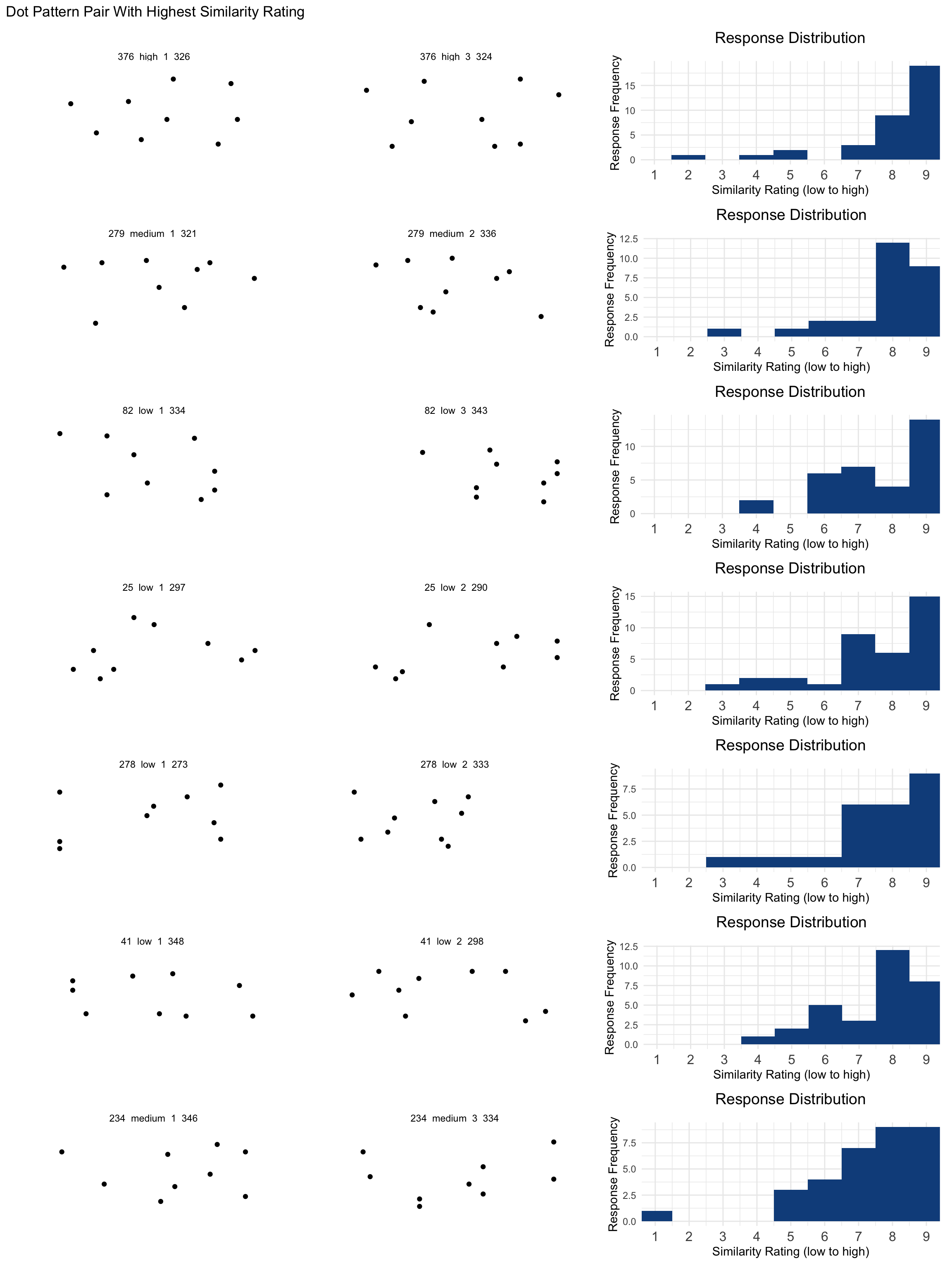
## Highest Rated Items
```{r}
#| fig-width: 12
#| fig-height: 16
# pairCounts |>
# slice_tail(n = 2) |>
# arrange(desc(mean_resp)) |>
# pull(pair_label) |>
# map(~plot_hist_pair(.x ))
ph <- pairCounts |>
slice_tail(n = 7) |>
arrange(desc(mean_resp)) |>
pull(pair_label) |>
map(~pair_hist_plot(.x, item_labels = TRUE)) |>
wrap_plots(ncol = 1) + plot_annotation(title = "Dot Pattern Pair With Highest Similarity Rating")
ph
```
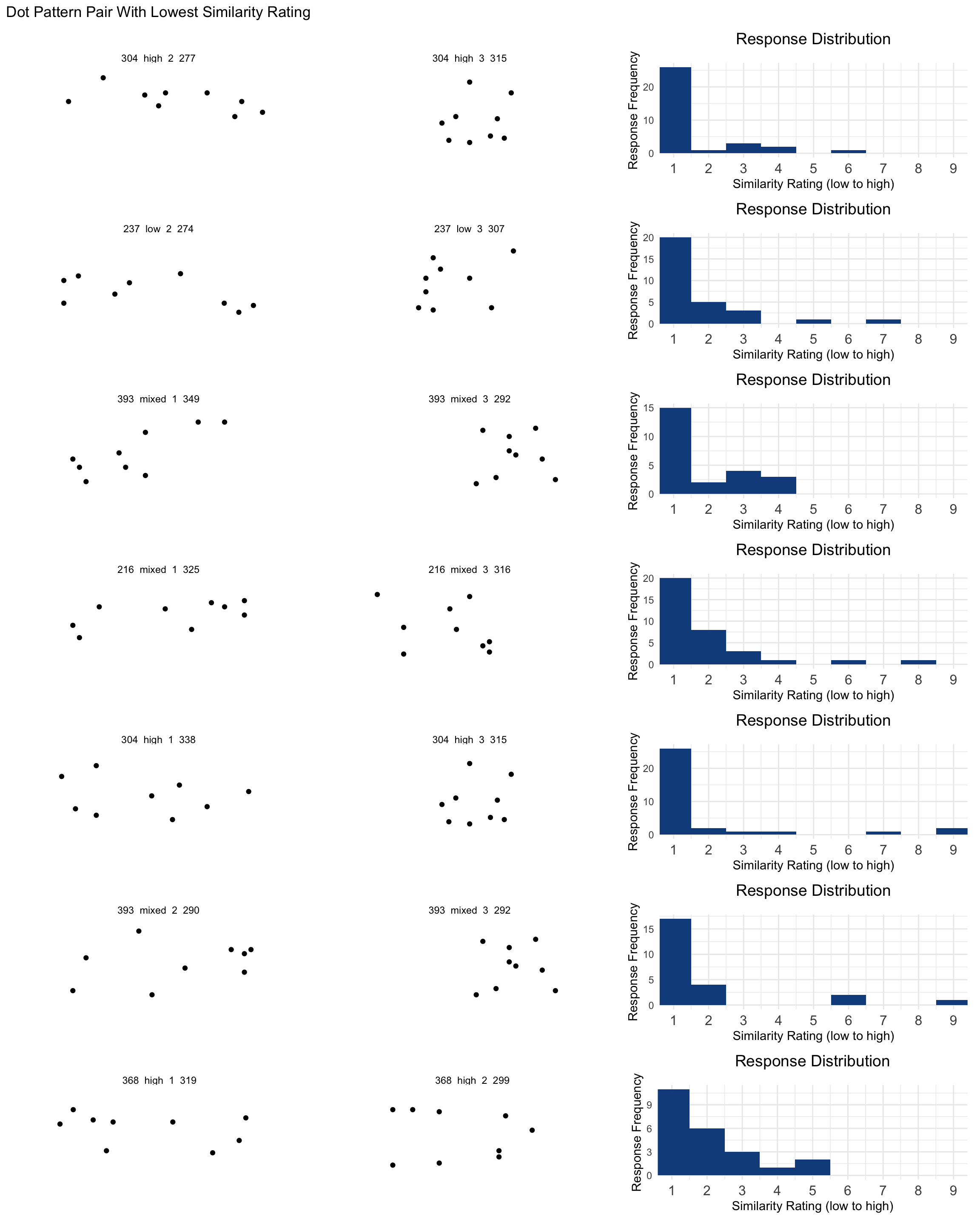
## Lowest Rated Items
```{r}
#| fig-width: 12
#| fig-height: 15
# pairCounts |>
# slice_tail(n = 2) |>
# arrange(desc(mean_resp)) |>
# pull(pair_label) |>
# map(~plot_hist_pair(.x ))
pl <- pairCounts |>
slice_head(n = 7) |>
pull(pair_label) |>
map(~pair_hist_plot(.x, item_labels = TRUE)) |>
wrap_plots(ncol = 1) + plot_annotation(title = "Dot Pattern Pair With Lowest Similarity Rating")
pl
```