Hu & Nosofsky 2024
Procedure
- Between Groups Training Manipulation
- low distortion - 4 posner levels from prototype
- medium distortion - 6 posner levels from prototype
- high distortion - 7.7 posner levels from prototype
- mixed distortion - equal number of low,medium, high patterns.
- 270 training trials (10 blocks of 27)
- training items are never repeated
- 84 Testing trials
- 27 old patterns (trained patterns) - at least 2 from each training block
- 3 from each prototype (1 per category)
- 9 novel low distortions
- 18 novel medium distortions
- 27 new high distortions
- testing items are never repeated
Hu & Nosofsky 2024
Training Stage
- Participants: 304 students from Indiana University.
- Conditions: Four training conditions (low-distortion, medium-distortion, high-distortion, mixed-distortion).
- Training Blocks: 10 blocks, each with 27 trials (270 trials total).
- Training Patterns: Different set of training patterns randomly generated in each block.
- Feedback: Corrective feedback provided for 2 seconds after each response.
- Training Instances: No individual training item was ever repeated.
Testing Stage
-
Test Patterns: 84 test trials including:
- 27 old patterns from the training phase.
- 3 prototypes (one per category).
- 9 new low-level distortions (three per category).
- 18 new medium-level distortions (six per category).
- 27 new high-level distortions (nine per category).
- Procedure: Each pattern presented once in a random order.
- Response Requirement: Participants classified all test patterns into one of the three candidate categories (no “None” response allowed).
Procedure for Creating Different Distortion Levels
- Prototype Generation: For each subject, prototypes for three categories generated by placing nine dots at random grid positions in a 30x30 area of a 50x50 grid.
-
Distortion Levels:
- Low Distortion: Dots displaced by an average of 4 Posner-levels.
- Medium Distortion: Dots displaced by an average of 6 Posner-levels.
- High Distortion: Dots displaced by an average of 7.7 Posner-levels.
- Pattern Generation: Each pattern constructed by displacing each dot by a random direction and distance according to the specified distortion level.
Testing
Filter to only include sbjs. who learned during training
Display code
# dCat |> filter(Phase==2) |> group_by(condit) |> summarise(n=n_distinct(sbjCode))
# dCat |> filter(finalTrain>.33, Phase==2) |> group_by(condit) |> summarise(n=n_distinct(sbjCode))
# dCat |> filter(finalTrain>.66, Phase==2) |> group_by(condit) |> summarise(n=n_distinct(sbjCode))
# dCat |> filter(finalTrain>.70, Phase==2) |> group_by(condit) |> summarise(n=n_distinct(sbjCode))
dCat |>
filter(Phase == 2) |>
group_by(condit) |>
summarise(
`All Sbjs.` = n_distinct(sbjCode),
`>.33` = n_distinct(sbjCode[finalTrain > .35]),
`>.50` = n_distinct(sbjCode[finalTrain > .50]),
`>.70` = n_distinct(sbjCode[finalTrain > .70])
) |> kable()| condit | All Sbjs. | >.33 | >.50 | >.70 |
|---|---|---|---|---|
| low | 77 | 77 | 75 | 73 |
| medium | 78 | 75 | 63 | 42 |
| mixed | 74 | 67 | 57 | 42 |
| high | 75 | 56 | 35 | 17 |
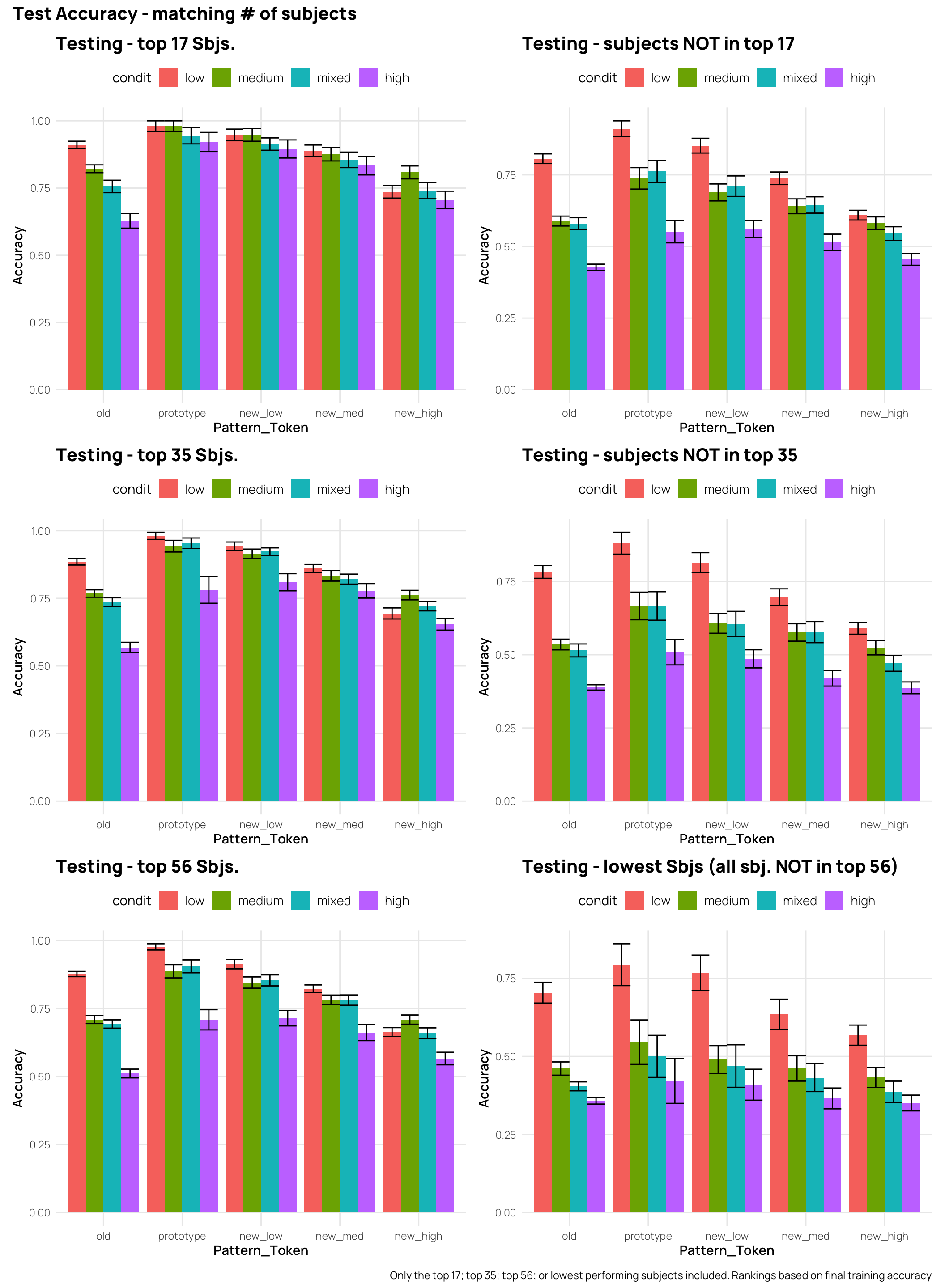
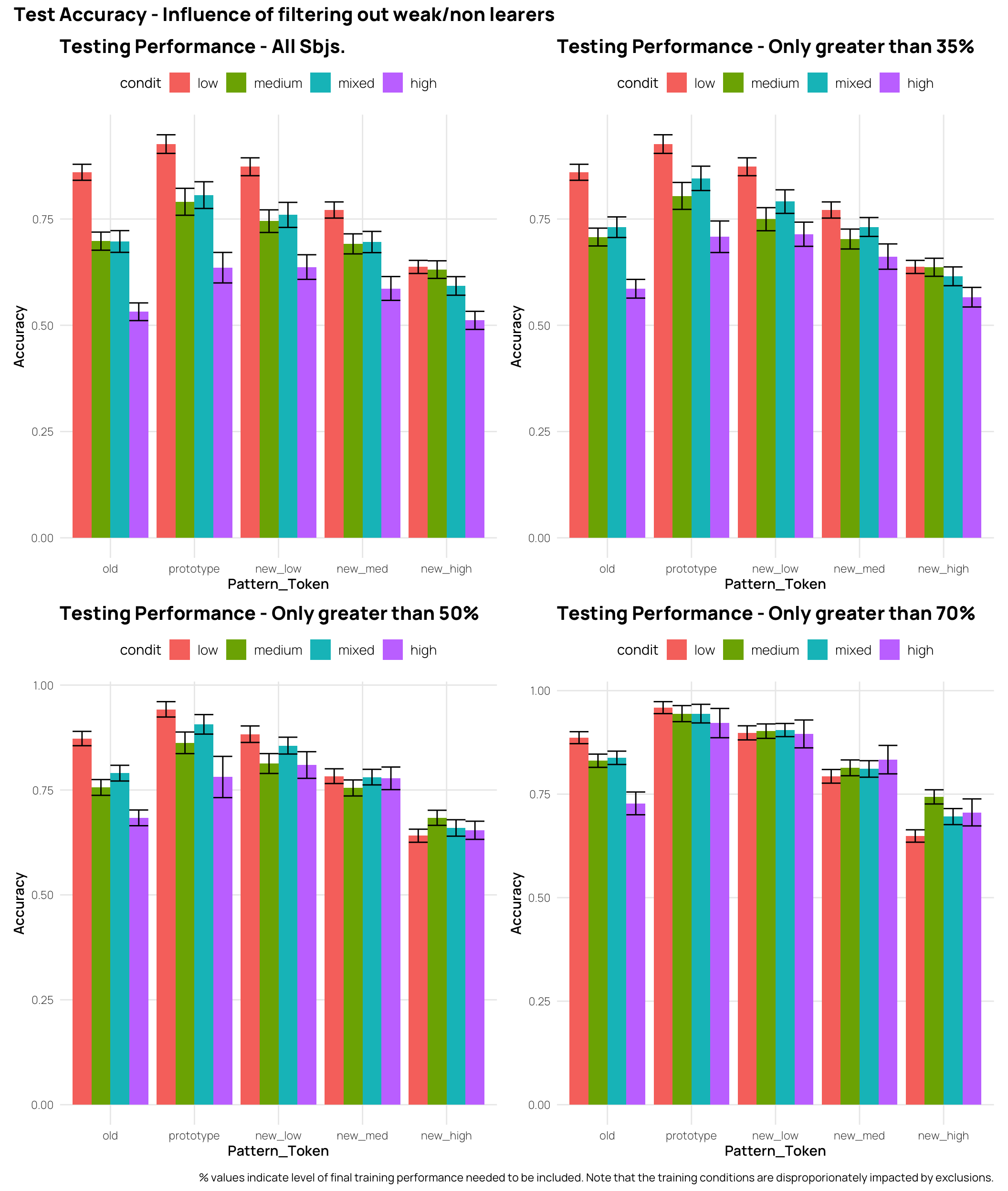
In the full data-set, the high distortion group has the worst performance for all testing patterns, and the low distortion group has performance either better or equal to all other training groups. However if we only include participants who exceeded 50%, or 70% accuracy during training - the patterns become a bit more complex. Considering the new_high distortion testing items, the groups that experienced more training variability now either match or outperform the low distortion group. The effect of filtering out the weaker learners does not influence the ordering of performance for the old items (i.e. The low distortion group remains the best, and the high distortion group remains the worst).
Display code
trainRanks <- dCat |> group_by(sbjCode,condit) |>
select(finalTrain) |> slice(1) |> arrange(-finalTrain)
top17 <- trainRanks |> group_by(condit) |> slice(1:17)
top35 <- trainRanks |> group_by(condit) |> slice(1:35)
top56 <- trainRanks |> group_by(condit) |> slice(1:56)
low <- trainRanks |> filter(!(sbjCode %in% top56$sbjCode))
#top17 |> gt::gt()
t17 <- dCat |> filter(sbjCode %in% top17$sbjCode) |>
group_by(sbjCode, condit, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=condit, group=condit)) +
stat_summary(geom="bar",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge()) +
labs(title="Testing - top 17 Sbjs.", y="Accuracy")
t35 <- dCat |> filter(sbjCode %in% top35$sbjCode) |>
group_by(sbjCode, condit, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=condit, group=condit)) +
stat_summary(geom="bar",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge()) +
labs(title="Testing - top 35 Sbjs.", y="Accuracy")
t56 <- dCat |> filter(sbjCode %in% top56$sbjCode) |>
group_by(sbjCode, condit, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=condit, group=condit)) +
stat_summary(geom="bar",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge()) +
labs(title="Testing - top 56 Sbjs.", y="Accuracy")
tLow56 <- dCat |> filter(sbjCode %in% low$sbjCode) |>
group_by(sbjCode, condit, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=condit, group=condit)) +
stat_summary(geom="bar",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge()) +
labs(title="Testing - lowest Sbjs (all sbj. NOT in top 56)", y="Accuracy")
tLow35 <- dCat |> filter(!(sbjCode %in% (trainRanks |> group_by(condit) |> slice(1:35))$sbjCode)) |>
group_by(sbjCode, condit, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=condit, group=condit)) +
stat_summary(geom="bar",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge()) +
labs(title="Testing - subjects NOT in top 35", y="Accuracy")
tLow17 <- dCat |> filter(!(sbjCode %in% (trainRanks |> group_by(condit) |> slice(1:17))$sbjCode)) |>
group_by(sbjCode, condit, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=condit, group=condit)) +
stat_summary(geom="bar",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge()) +
labs(title="Testing - subjects NOT in top 17", y="Accuracy")
(t17+tLow17) /(t35+tLow35)/(t56+tLow56) +
plot_annotation(title="Test Accuracy - matching # of subjects",
caption=" Only the top 17; top 35; top 56; or lowest performing subjects included. Rankings based on final training accuracy")Display code
# (t17+t35) /(t56+tLow) +
# plot_annotation(title="Test Accuracy - matching # of subjects",
# caption=" Only the top 17; top 35; top 56; or lowest performing subjects included. Rankings based on final training accuracy")Display code
tAll <- dCat |> filter(Phase==2) |>
group_by(sbjCode, condit, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=condit, group=condit)) +
stat_summary(geom="bar",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge()) +
labs(title="Testing Performance - All Sbjs.", y="Accuracy")
t33 <- dCat |> filter(finalTrain>.35, Phase==2) |>
group_by(sbjCode, condit, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=condit, group=condit)) +
stat_summary(geom="bar",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge()) +
labs(title="Testing Performance - Only greater than 35%", y="Accuracy")
t66 <- dCat |> filter(finalTrain>.50, Phase==2) |>
group_by(sbjCode, condit,Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=condit, group=condit)) +
stat_summary(geom="bar",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge()) +
labs(title="Testing Performance - Only greater than 50%", y="Accuracy")
t80 <- dCat |> filter(finalTrain>.70, Phase==2) |>
group_by(sbjCode, condit, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=condit, group=condit)) +
stat_summary(geom="bar",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge()) +
labs(title="Testing Performance - Only greater than 70%", y="Accuracy")
((tAll + t33)/(t66 + t80)) +
plot_annotation(title="Test Accuracy - Influence of filtering out weak/non learers",
caption=" % values indicate level of final training performance needed to be included. Note that the training conditions are disproporionately impacted by exclusions.")Display code
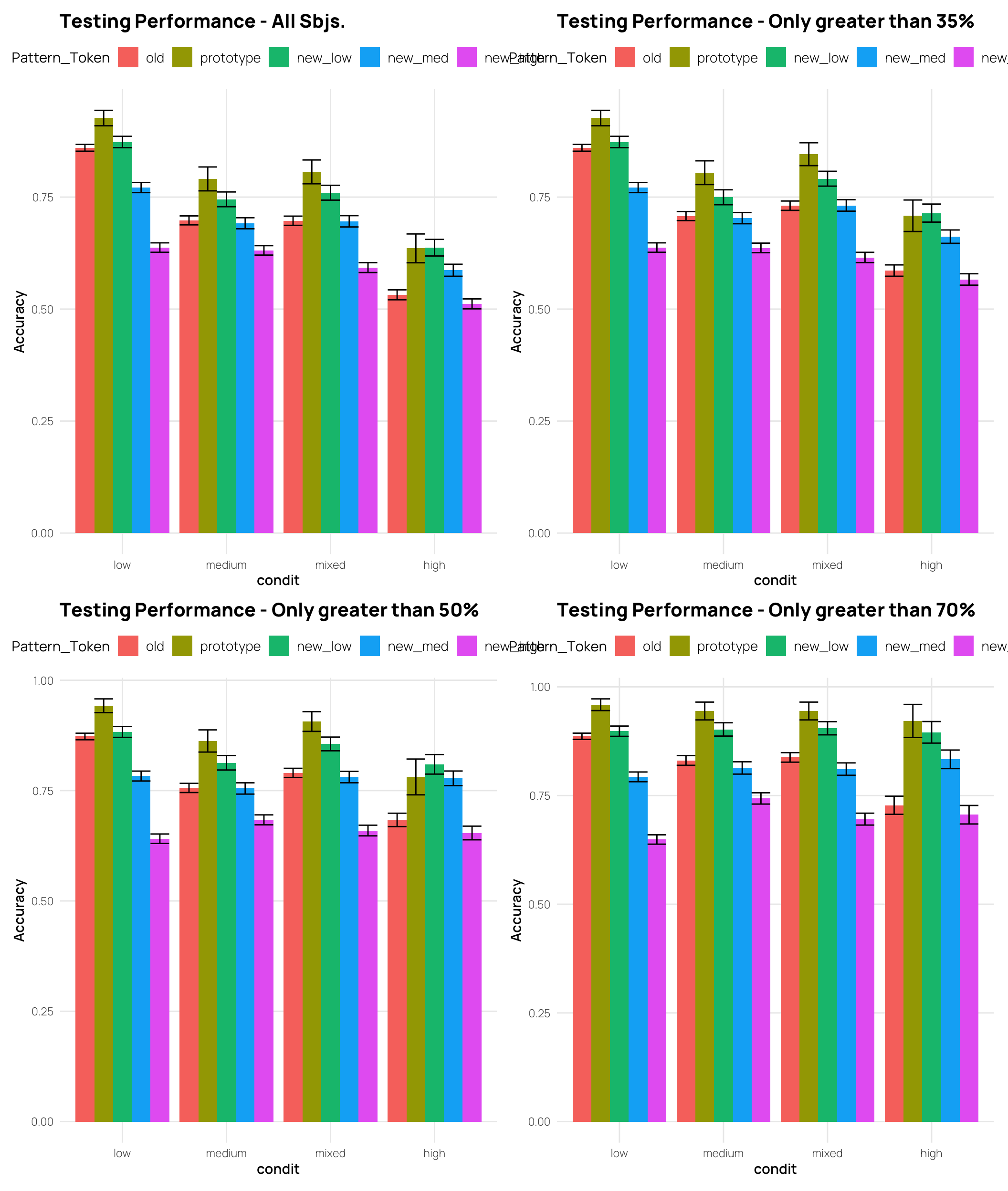
tAll <- dCat |> filter(Phase==2) |>
ggplot(aes(x=condit, y=Corr, fill=Pattern_Token, group=Pattern_Token)) +
stat_summary(geom="bar",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge()) +
labs(title="Testing Performance - All Sbjs.", y="Accuracy")
t33 <- dCat |> filter(finalTrain>.35, Phase==2) |>
ggplot(aes(x=condit, y=Corr, fill=Pattern_Token, group=Pattern_Token)) +
stat_summary(geom="bar",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge()) +
labs(title="Testing Performance - Only greater than 35%", y="Accuracy")
t66 <- dCat |> filter(finalTrain>.50, Phase==2) |>
ggplot(aes(x=condit, y=Corr, fill=Pattern_Token, group=Pattern_Token)) +
stat_summary(geom="bar",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge()) +
labs(title="Testing Performance - Only greater than 50%", y="Accuracy")
t80 <- dCat |> filter(finalTrain>.70, Phase==2) |>
ggplot(aes(x=condit, y=Corr, fill=Pattern_Token, group=Pattern_Token)) +
stat_summary(geom="bar",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge()) +
labs(title="Testing Performance - Only greater than 70%", y="Accuracy")
((tAll + t33)/(t66 + t80))Split by Quartiles (end of training performance)
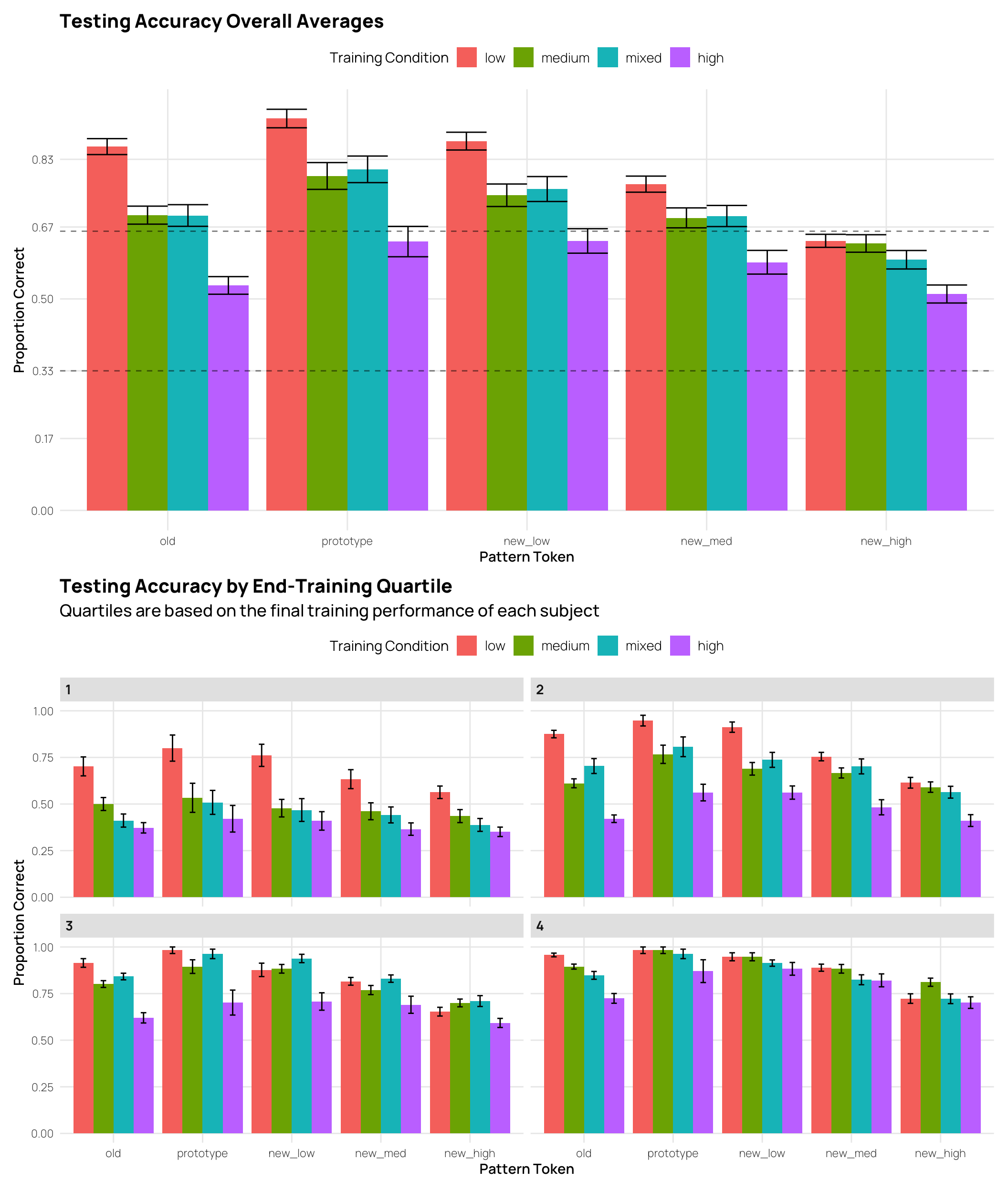
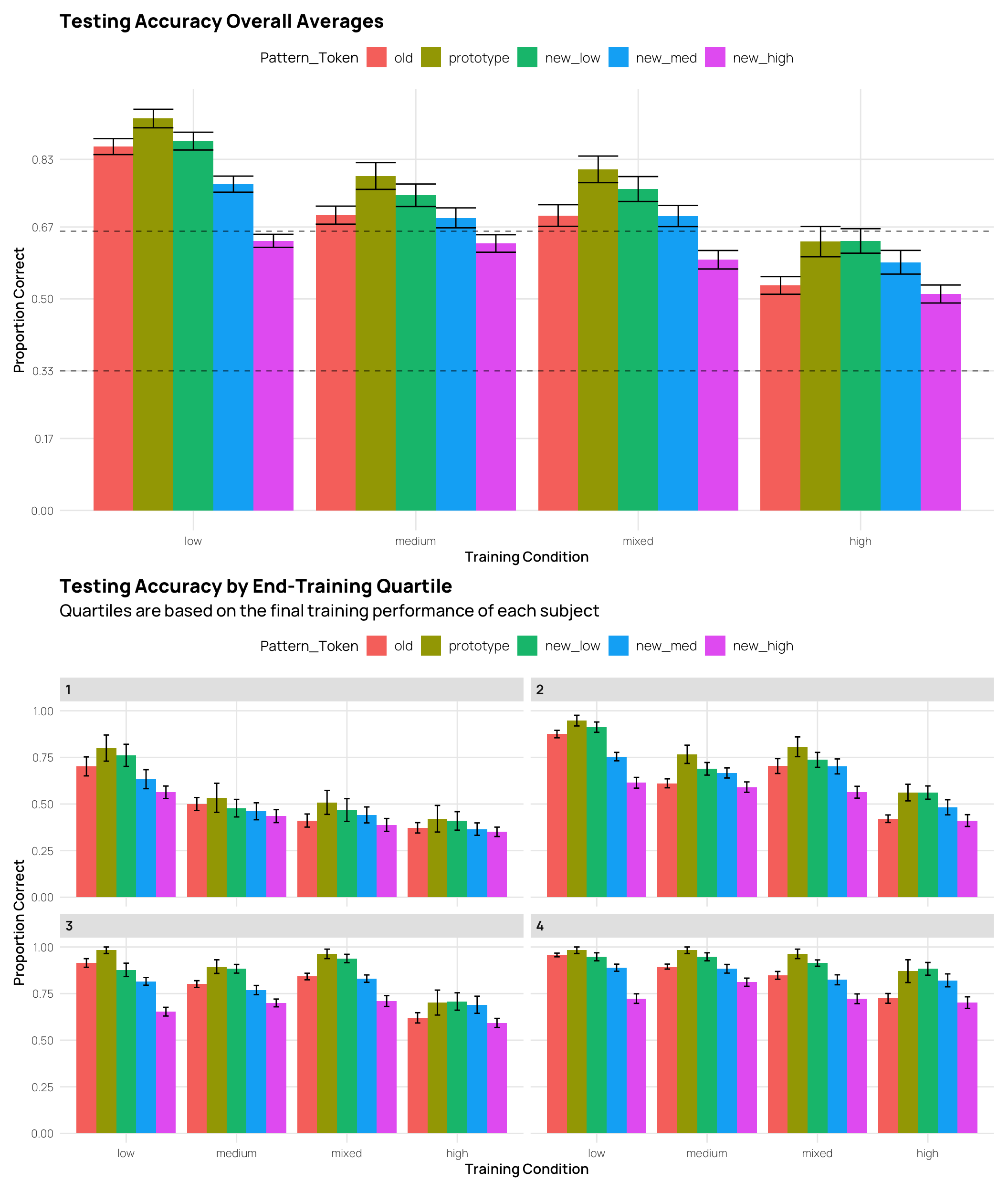
We can also inspect testing performance by splitting the data into quartiles based on the final training performance. This avoids the issue of excluding subjects, but increases the disparity in training performance between groups (i.e. the worst quartile of high distortion sbjs. had much worse training performance than the worst quartile of low distortion sbjs.)
Display code
tx1 <- theme(axis.title.x=element_blank(), axis.text.x=element_blank())
tx2 <- theme(axis.title.x=element_blank(), axis.text.x=element_blank(),legend.position = "none" )
yt <- round(seq(0,1,length.out=7), 2)
eg <- list(geom_hline(yintercept = c(.33, .66),linetype="dashed", alpha=.5),scale_y_continuous(breaks=yt))
dq1 <- dCat |> filter(Phase==2) |>
group_by(sbjCode, condit, quartile, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=condit)) +
stat_summary(geom="bar",fun="mean", position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge(), width=.9) +
eg + labs(x="Pattern Token", y="Proportion Correct", title="Testing Accuracy Overall Averages",
fill="Training Condition")
dq2 <-dCat |> filter(Phase == 2) |>
group_by(sbjCode, condit, quartile, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x = Pattern_Token, y = Corr, fill = condit)) +
stat_summary(geom = "bar", fun = "mean", position = position_dodge()) +
stat_summary(geom = "errorbar", fun.data = mean_se, position = position_dodge(width = 0.9), width = 0.25) +
facet_wrap(~quartile) +
labs(x = "Pattern Token", y = "Proportion Correct", title = "Testing Accuracy by End-Training Quartile",
subtitle="Quartiles are based on the final training performance of each subject",
fill="Training Condition")
dq1/dq2Display code
tx1 <- theme(axis.title.x=element_blank(), axis.text.x=element_blank())
tx2 <- theme(axis.title.x=element_blank(), axis.text.x=element_blank(),legend.position = "none" )
yt <- round(seq(0,1,length.out=7), 2)
eg <- list(geom_hline(yintercept = c(.33, .66),linetype="dashed", alpha=.5),scale_y_continuous(breaks=yt))
dq1 <- dCat |> filter(Phase==2) |>
group_by(sbjCode, condit, quartile, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x=condit, y=Corr, fill=Pattern_Token)) +
stat_summary(geom="bar",fun="mean", position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge(), width=.9) +
eg + labs(x="Training Condition", y="Proportion Correct", title="Testing Accuracy Overall Averages")
dq2 <-dCat |> filter(Phase == 2) |>
group_by(sbjCode, condit, quartile, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x = condit, y = Corr, fill = Pattern_Token)) +
stat_summary(geom = "bar", fun = "mean", position = position_dodge()) +
stat_summary(geom = "errorbar", fun.data = mean_se, position = position_dodge(width = 0.9), width = 0.25) +
facet_wrap(~quartile) +
labs(x = "Training Condition", y = "Proportion Correct", title = "Testing Accuracy by End-Training Quartile",
subtitle="Quartiles are based on the final training performance of each subject")
dq1/dq2Display code
dCat |> filter(Phase == 2) |>
group_by(sbjCode, condit, quartile, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x = condit, y = Corr, fill = Pattern_Token)) +
geom_boxplot(position=position_dodge()) +
geom_jitter(position = position_jitterdodge(jitter.width = 0.25, dodge.width = 0.9), alpha = .2) +
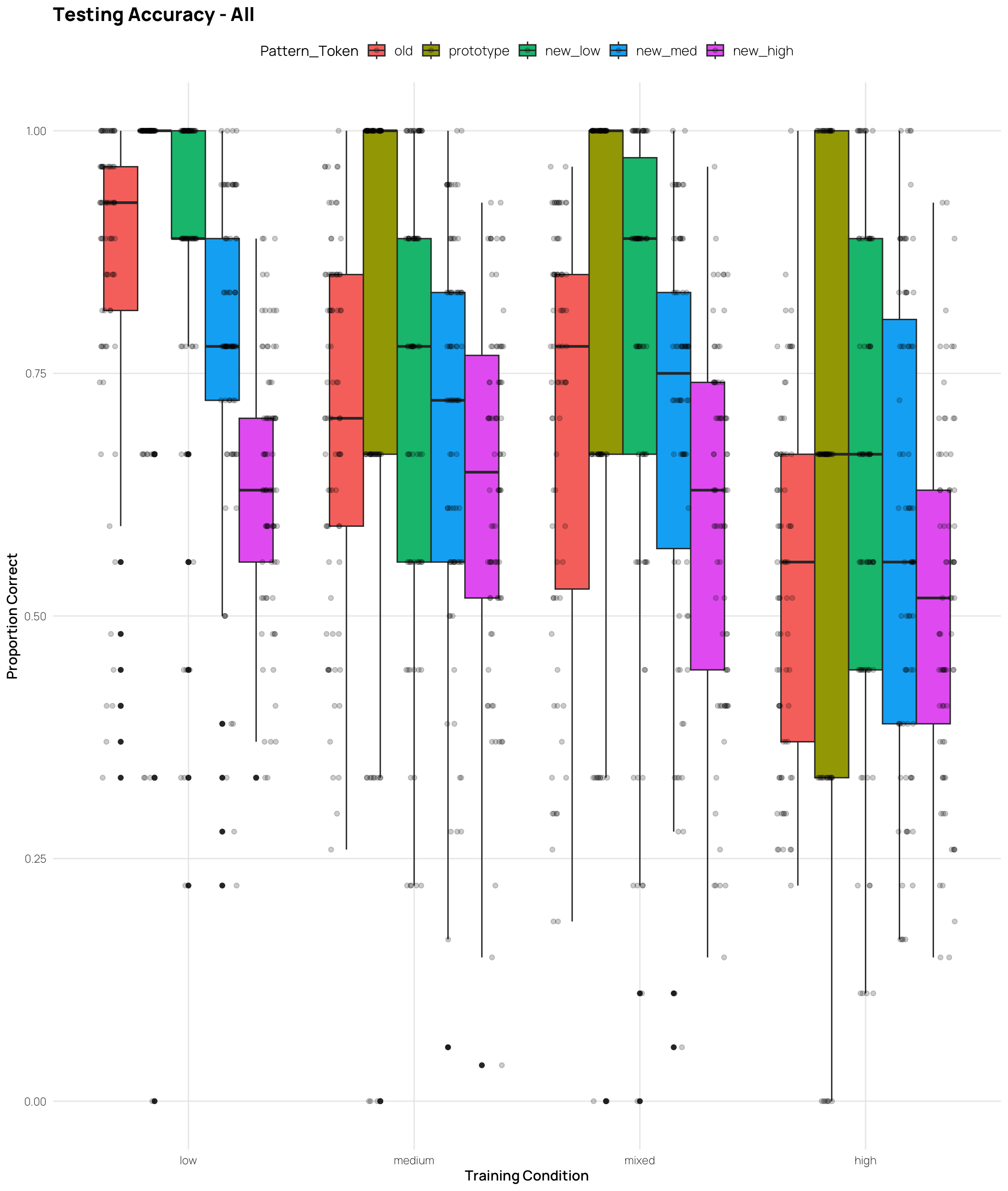
labs(x = "Training Condition", y = "Proportion Correct", title = "Testing Accuracy - All")Display code
dCat |> filter(Phase == 2) |>
group_by(sbjCode, condit, quartile, Pattern_Token) |>
summarize(Corr=mean(Corr)) |>
ggplot(aes(x = condit, y = Corr, fill = Pattern_Token)) +
geom_boxplot(position=position_dodge()) +
geom_jitter(position = position_jitterdodge(jitter.width = 0.25, dodge.width = 0.9), alpha = .2) +
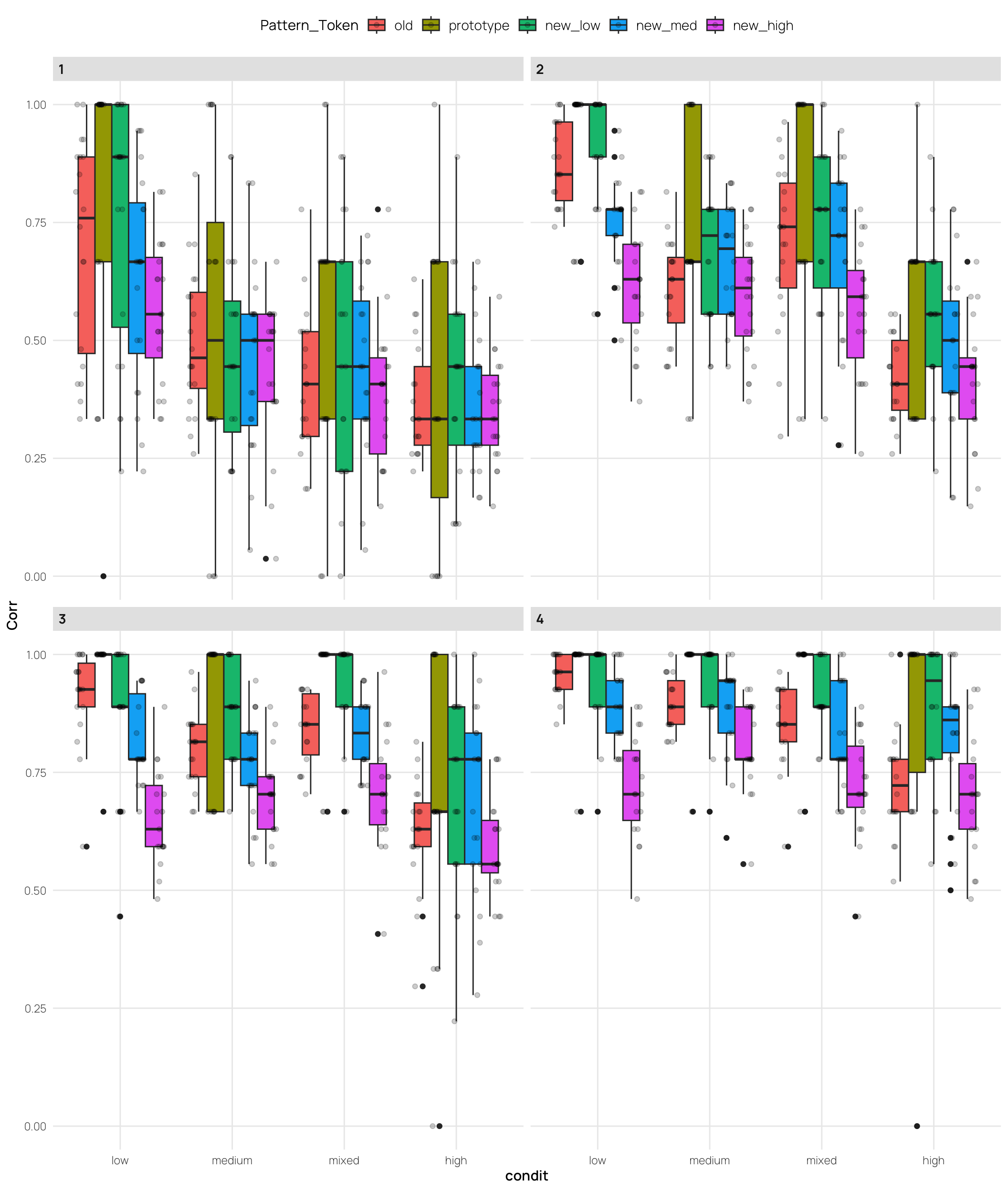
facet_wrap(~quartile)Display code
labs(x = "Training Condition", y = "Proportion Correct", title = "Testing Accuracy - All")$x
[1] "Training Condition"
$y
[1] "Proportion Correct"
$title
[1] "Testing Accuracy - All"
attr(,"class")
[1] "labels"Testing Reaction Time
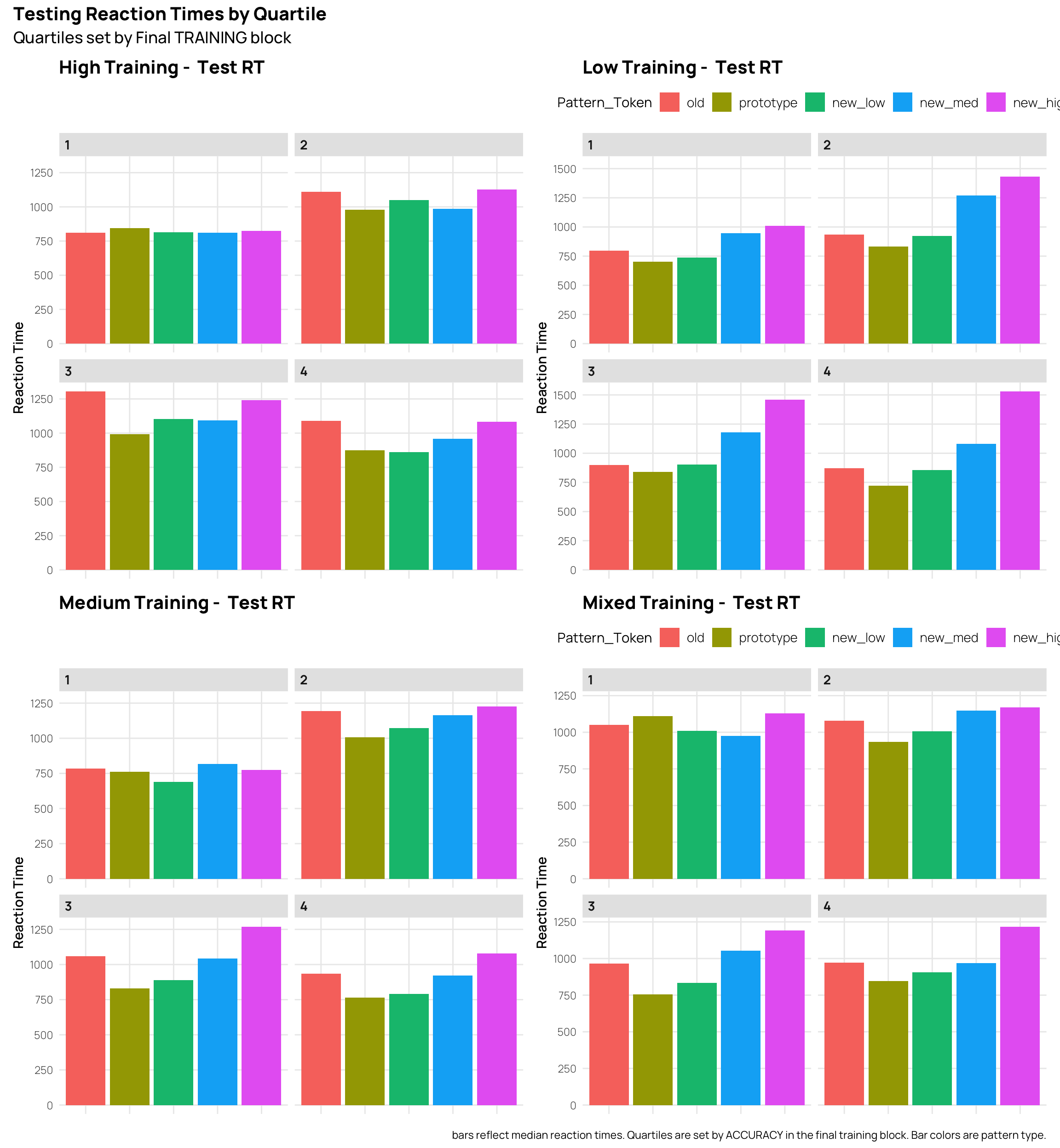
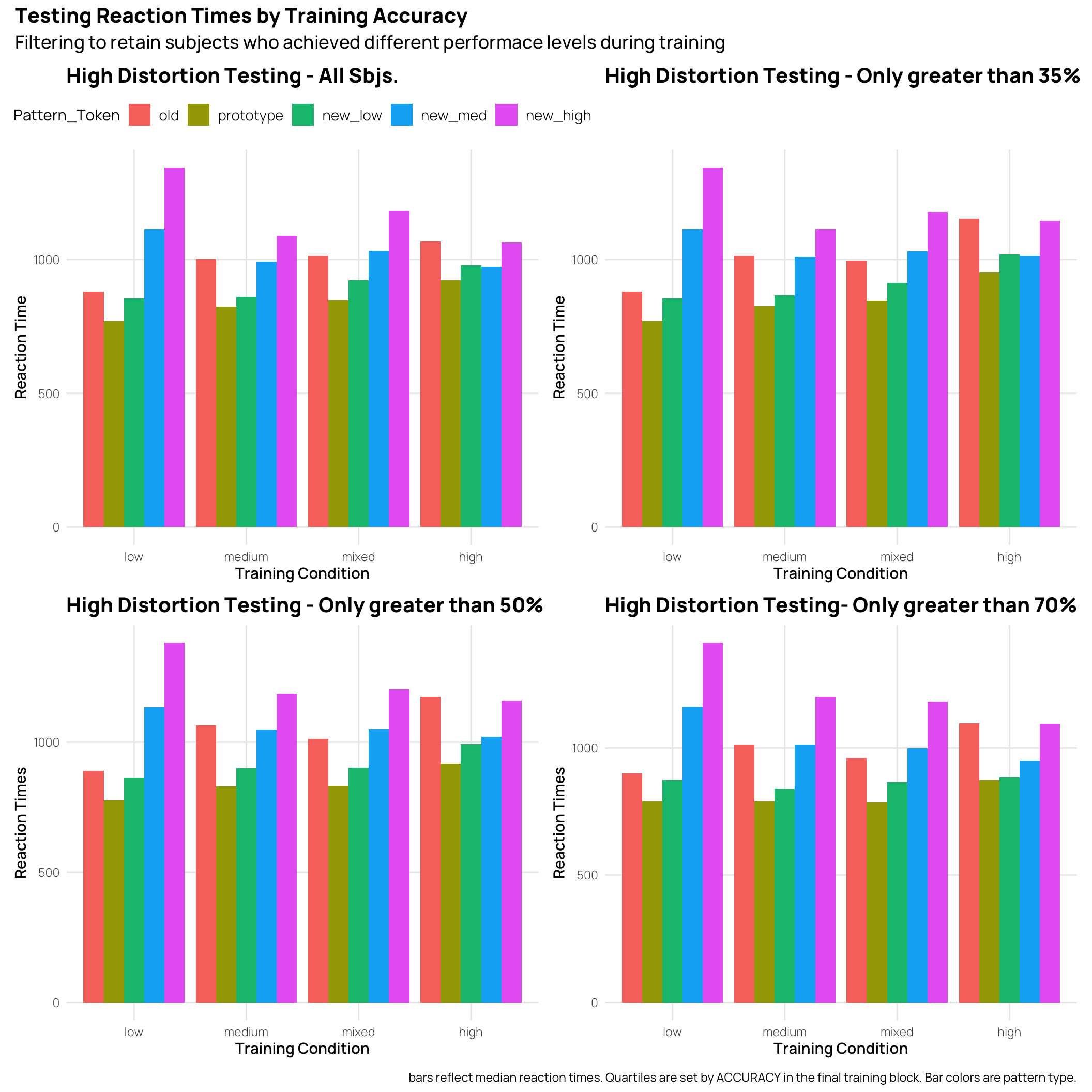
Worth comparing the RT’s to the accuracy. In many cases the RT’s show the inverse pattern of accuracy, i.e. slower RT’s for less accurate patterns.But, the weakest quartile for the High and Medium distortion training conditions don’t follow this pattern.
Display code
tx1 <- theme(axis.title.x=element_blank(), axis.text.x=element_blank())
tx2 <- theme(axis.title.x=element_blank(), axis.text.x=element_blank(),legend.position = "none" )
rtfun <- "median"
yt <- round(seq(0,1500,length.out=7), 2)
eg <- list(scale_y_continuous(breaks=yt))
htq <- dCat |> filter(condit=="high", Phase==2) |>
ggplot(aes(x=Pattern_Token, y=rt, fill=Pattern_Token)) +
stat_summary(geom="bar",fun=rtfun)+
facet_wrap(~quartile) + eg+
labs(title="High Training - Test RT", y="Reaction Time") +tx2
ltq <- dCat |> filter(condit=="low", Phase==2) |>
ggplot(aes(x=Pattern_Token, y=rt, fill=Pattern_Token)) +
stat_summary(geom="bar",fun=rtfun)+
facet_wrap(~quartile) + eg+
labs(title="Low Training - Test RT", y="Reaction Time") +tx1
mtq <- dCat |> filter(condit=="medium", Phase==2) |>
ggplot(aes(x=Pattern_Token, y=rt, fill=Pattern_Token)) +
stat_summary(geom="bar",fun=rtfun)+
facet_wrap(~quartile) + eg+
labs(title="Medium Training - Test RT", y="Reaction Time") +tx2
mxtq <- dCat |> filter(condit=="mixed", Phase==2) |>
ggplot(aes(x=Pattern_Token, y=rt, fill=Pattern_Token)) +
stat_summary(geom="bar",fun=rtfun)+
facet_wrap(~quartile) + eg+
labs(title="Mixed Training - Test RT", y="Reaction Time") + tx1
(htq+ltq)/(mtq+mxtq) + plot_annotation(
title = 'Testing Reaction Times by Quartile',
subtitle = 'Quartiles set by Final TRAINING block',
caption = 'bars reflect median reaction times. Quartiles are set by ACCURACY in the final training block. Bar colors are pattern type.'
)Display code
tAll <- dCat |> filter(Phase==2) |>
ggplot(aes(x=condit, y=rt, fill=Pattern_Token, group=Pattern_Token)) +
stat_summary(geom="bar",fun=rtfun, position=position_dodge())+
labs(title="High Distortion Testing - All Sbjs.", y="Reaction Time", x="Training Condition") + theme(legend.position = "top")
t33 <- dCat |> filter(finalTrain>.35, Phase==2) |>
ggplot(aes(x=condit, y=rt, fill=Pattern_Token, group=Pattern_Token)) +
stat_summary(geom="bar",fun=rtfun, position=position_dodge())+
labs(title="High Distortion Testing - Only greater than 35%", y="Reaction Time", x="Training Condition") + theme(legend.position = "none")
t66 <- dCat |> filter(finalTrain>.50, Phase==2) |>
ggplot(aes(x=condit, y=rt, fill=Pattern_Token, group=Pattern_Token)) +
stat_summary(geom="bar",fun=rtfun, position=position_dodge())+
labs(title="High Distortion Testing - Only greater than 50%", y="Reaction Times", x="Training Condition") + theme(legend.position = "none")
t80 <- dCat |> filter(finalTrain>.70, Phase==2) |>
ggplot(aes(x=condit, y=rt, fill=Pattern_Token, group=Pattern_Token)) +
stat_summary(geom="bar",fun=rtfun, position=position_dodge())+
labs(title="High Distortion Testing- Only greater than 70%", y="Reaction Times", x="Training Condition") + theme(legend.position = "none")
((tAll + t33)/(t66 + t80)) + plot_annotation(
title = 'Testing Reaction Times by Training Accuracy',
subtitle = 'Filtering to retain subjects who achieved different performace levels during training',
caption = 'bars reflect median reaction times. Quartiles are set by ACCURACY in the final training block. Bar colors are pattern type.'
)’
Training
Display code
tx1 <- theme(axis.title.x=element_blank(), axis.text.x=element_blank())
tx2 <- theme(axis.title.x=element_blank(), axis.text.x=element_blank(),legend.position = "none" )
yt <- round(seq(0,1,length.out=7), 2)
xt <- seq(1,10,1)
eg <- list(geom_hline(yintercept = c(.33, .66),linetype="dashed", alpha=.5),
scale_y_continuous(breaks=yt),
scale_x_continuous(breaks=xt))
tlt <- theme(legend.position = "top")
tln <- theme(legend.position = "none")
lavg <- dCat |> filter(Phase==1) |>
ggplot(aes(x=Block, y=Corr, col=condit, group=condit)) +
stat_summary(geom="line",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge(), width=.1) +
geom_smooth()+
labs(title="Average Learning Curves", y="Accuracy") + eg + tlt
lavgDist <- dCat |> filter(Phase==1) |>
group_by(sbjCode,condit, Block) |>
summarise(Corr=mean(Corr)) |>
ggplot(aes(x=Block, y=Corr, col=condit, group=condit)) +
ggdist::stat_pointinterval(alpha=.5, position=position_dodge()) +
#geom_smooth() +
labs(title="Average Learning Curves", y="Accuracy") + eg +tlt
lqt <- dCat |> filter(Phase==1) |>
ggplot(aes(x=Block, y=Corr, col=condit, group=condit)) +
stat_summary(geom="line",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge(), width=.1) +
facet_wrap(~quartile) +
labs(title="Learning Curves - End Training Quartiles",
subtitle=stringr::str_wrap("Quartiles are based on accuracy in the final training block (within condition)",65),
y="Accuracy") +
eg + tln
lqte <- dCat |> filter(Phase==1) |>
ggplot(aes(x=Block, y=Corr, col=condit, group=condit)) +
stat_summary(geom="line",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge(), width=.1) +
facet_wrap(~q_test_high) +
labs(title="Learning Curves - Test High Distortion Quartiles",
subtitle=stringr::str_wrap("Quartiles are based on accuracy on the new high distortion TEST items (within condition)",60),
y="Accuracy") +
eg+ tln
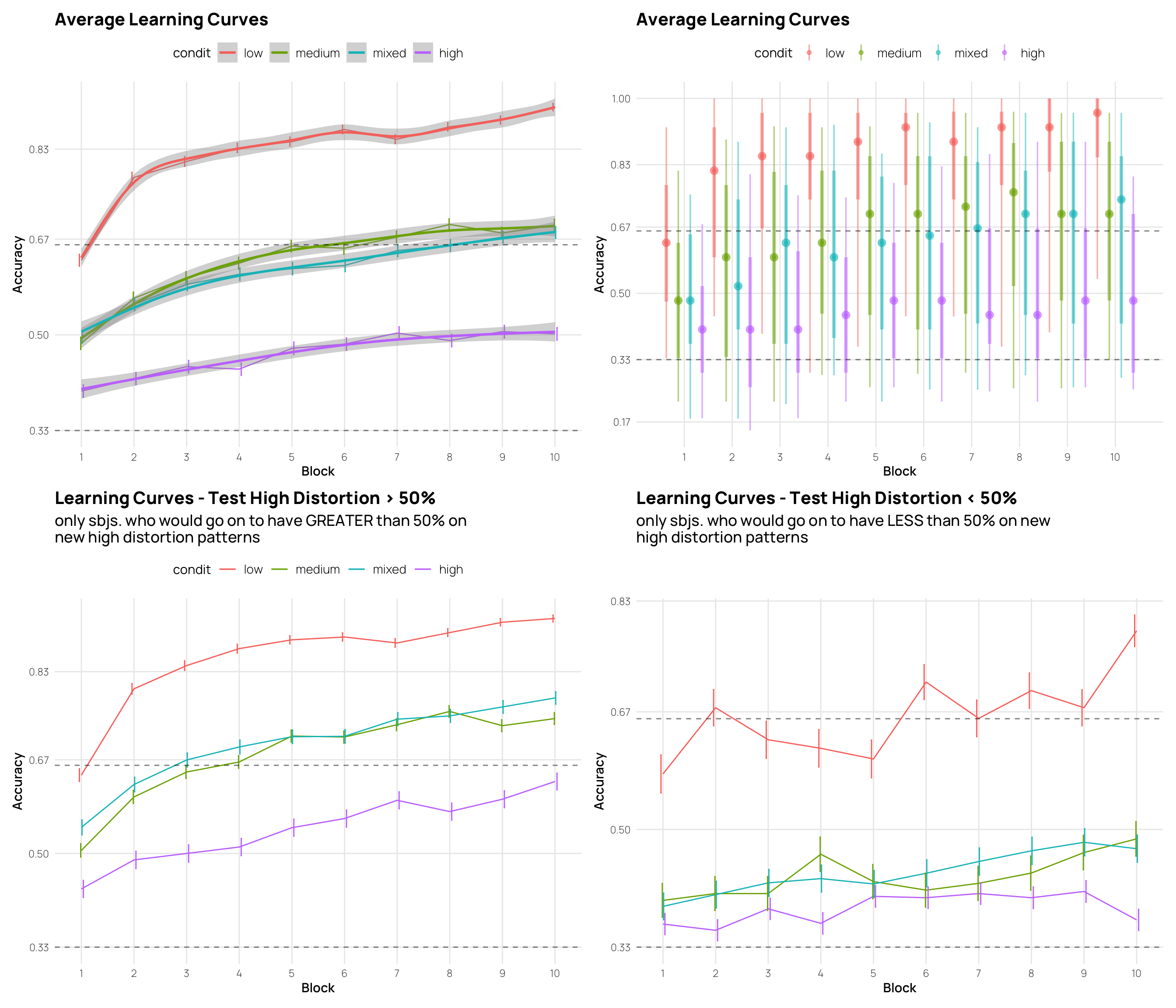
lte_g50h <- dCat |> filter(Phase==1, test_high>.50) |>
ggplot(aes(x=Block, y=Corr, col=condit, group=condit)) +
stat_summary(geom="line",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge(), width=.1) +
labs(title="Learning Curves - Test High Distortion > 50%",
subtitle=stringr::str_wrap("only sbjs. who would go on to have GREATER than 50% on new high distortion patterns",55),
y="Accuracy") +
eg+ tlt
lte_l50h <- dCat |> filter(Phase==1, test_high<.50) |>
ggplot(aes(x=Block, y=Corr, col=condit, group=condit)) +
stat_summary(geom="line",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge(), width=.1) +
labs(title="Learning Curves - Test High Distortion < 50%",
subtitle=stringr::str_wrap("only sbjs. who would go on to have LESS than 50% on new high distortion patterns",55),
y="Accuracy") +
eg+ tln
# lte_g50o <- dCat |> filter(Phase==1, test_low>.42) |>
# ggplot(aes(x=Block, y=Corr, col=condit, group=condit)) +
# stat_summary(geom="line",fun=mean, position=position_dodge())+
# stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge(), width=.1) +
# labs(title="Learning Curves - Test High Distortion > 50%",
# subtitle=stringr::str_wrap("only sbjs. who would go on to have GREATER than 50% on new high distortion patterns",55),
# y="Accuracy") +
# eg+ tlt
#
#
# lte_l50o <- dCat |> filter(Phase==1, test_low<.50) |>
# ggplot(aes(x=Block, y=Corr, col=condit, group=condit)) +
# stat_summary(geom="line",fun=mean, position=position_dodge())+
# stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge(), width=.1) +
# labs(title="Learning Curves - Test High Distortion < 50%",
# subtitle=stringr::str_wrap("only sbjs. who would go on to have LESS than 50% on new high distortion patterns",55),
# y="Accuracy") +
# eg+ tln
#((lavg +lavgDist)/(lqt + lqte) / (lte_g50 + lte_l50) / (lte_g70 + lte_l40))
#((lavg +lavgDist)/(lte_g50h + lte_l50h) / (lte_g50o + lte_l50o))
((lavg +lavgDist)/(lte_g50h + lte_l50h)) Display code
yt1 <- round(seq(500,4000,length.out=7), 2)
eg2 <- list(scale_y_continuous(breaks=yt, limits=c(min(yt1),max(yt1))))
eg1 <- list(scale_y_continuous(breaks=yt, n.breaks=7))
lavg <- dCat |> filter(Phase==1) |>
group_by(sbjCode,condit, Block) |>
summarise(rt=median(rt)) |>
ggplot(aes(x=Block, y=rt, col=condit, group=condit)) +
stat_summary(geom="line",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge(), width=.1) +
geom_smooth()+
labs(title="Average Learning Curves", y="Accuracy") +tlt
lavgDist <- dCat |> filter(Phase==1) |>
group_by(sbjCode,condit, Block) |>
summarise(rt=median(rt)) |>
ggplot(aes(x=Block, y=rt, col=condit, group=condit)) +
ggdist::stat_pointinterval(alpha=.5, position=position_dodge()) +
#geom_smooth() +
labs(title="Average Learning Curves", y="Accuracy") +tlt
lte_g50h <- dCat |> filter(Phase==1, test_high>.50) |>
group_by(sbjCode,condit, Block) |>
summarise(rt=median(rt)) |>
ggplot(aes(x=Block, y=rt, col=condit, group=condit)) +
stat_summary(geom="line",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge(), width=.1) +
labs(title="Learning Curves - Test High Distortion > 50%",
subtitle=stringr::str_wrap("only sbjs. who would go on to have GREATER than 50% on new high distortion patterns",55),
y="Accuracy") +
tlt
lte_l50h <- dCat |> filter(Phase==1, test_high<.50) |>
group_by(sbjCode,condit, Block) |>
summarise(rt=median(rt)) |>
ggplot(aes(x=Block, y=rt, col=condit, group=condit)) +
stat_summary(geom="line",fun=mean, position=position_dodge())+
stat_summary(geom="errorbar", fun.data=mean_se, position=position_dodge(), width=.1) +
labs(title="Learning Curves - Test High Distortion < 50%",
subtitle=stringr::str_wrap("only sbjs. who would go on to have LESS than 50% on new high distortion patterns",55),
y="Accuracy") +
tln
((lavg +lavgDist)/(lte_g50h + lte_l50h)) Individual Differences
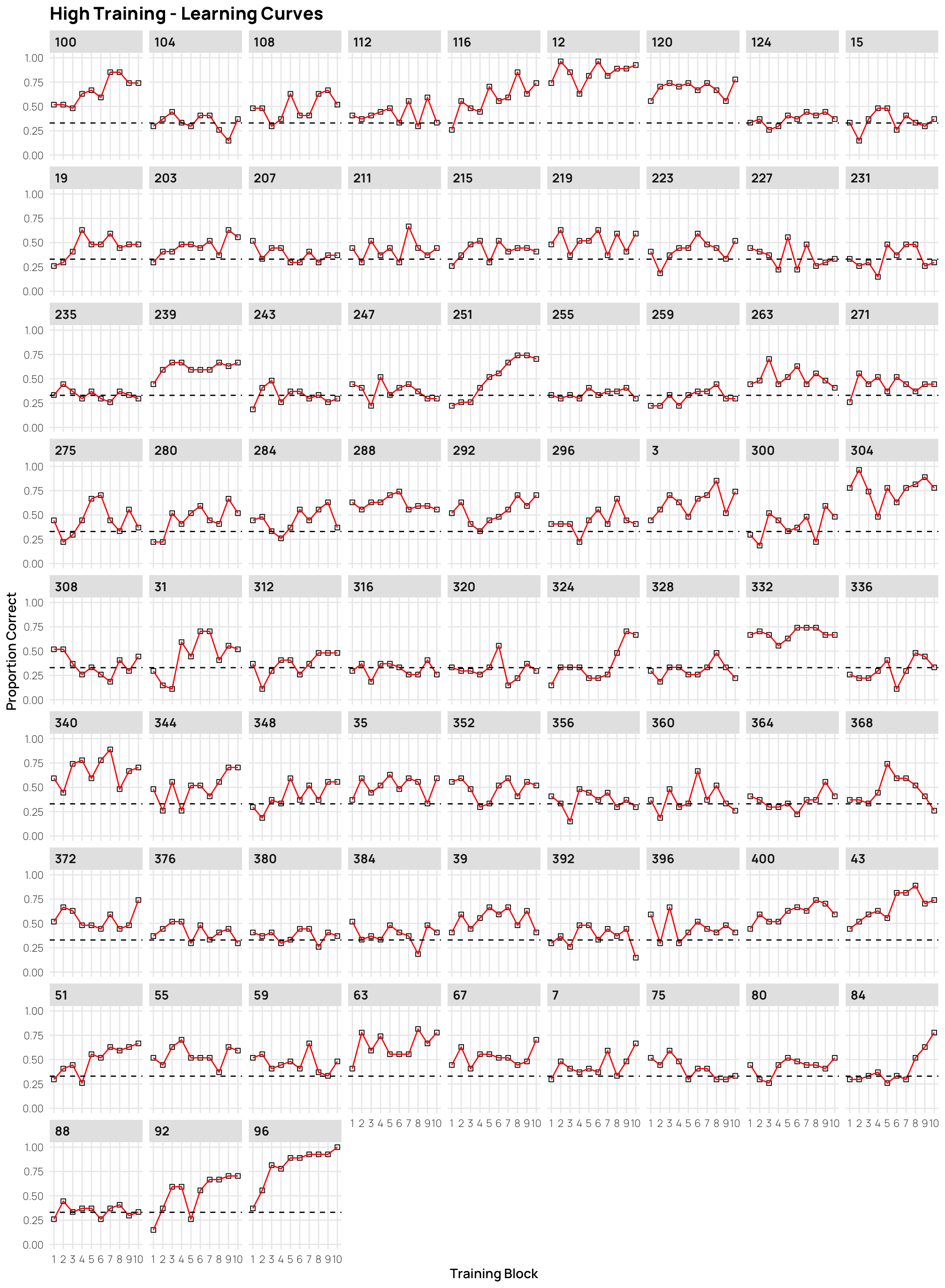
Individual Learning Curves
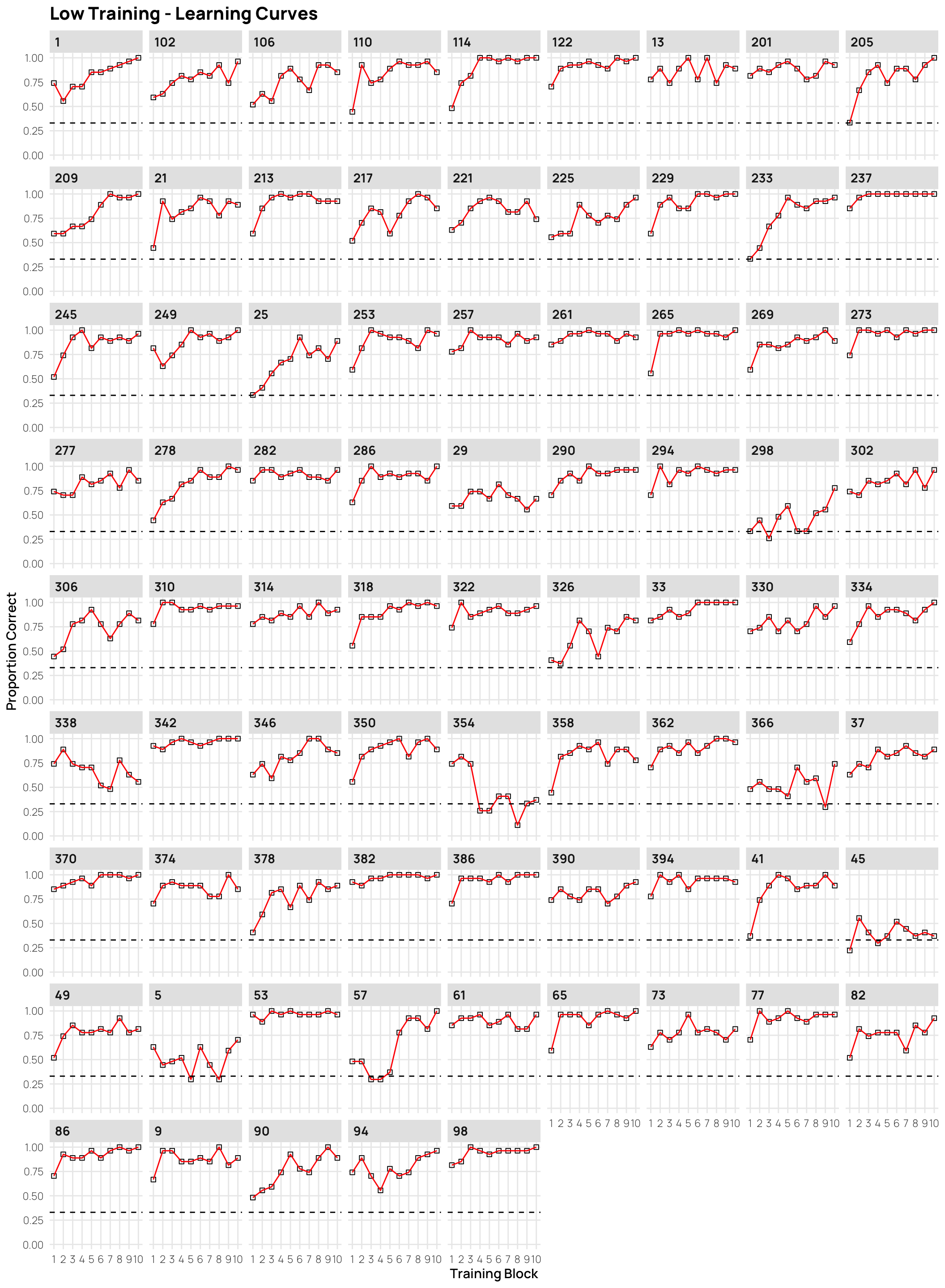
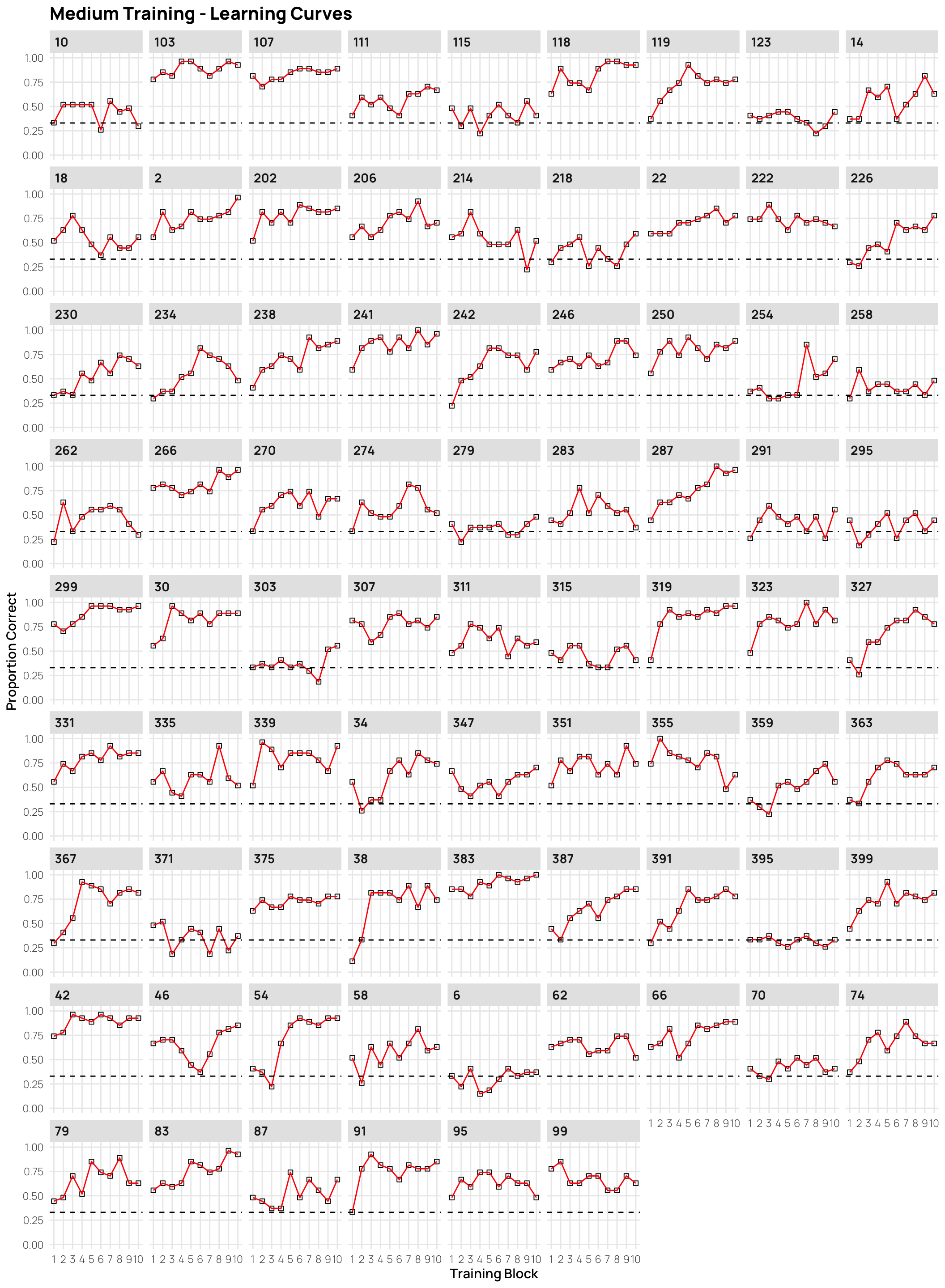
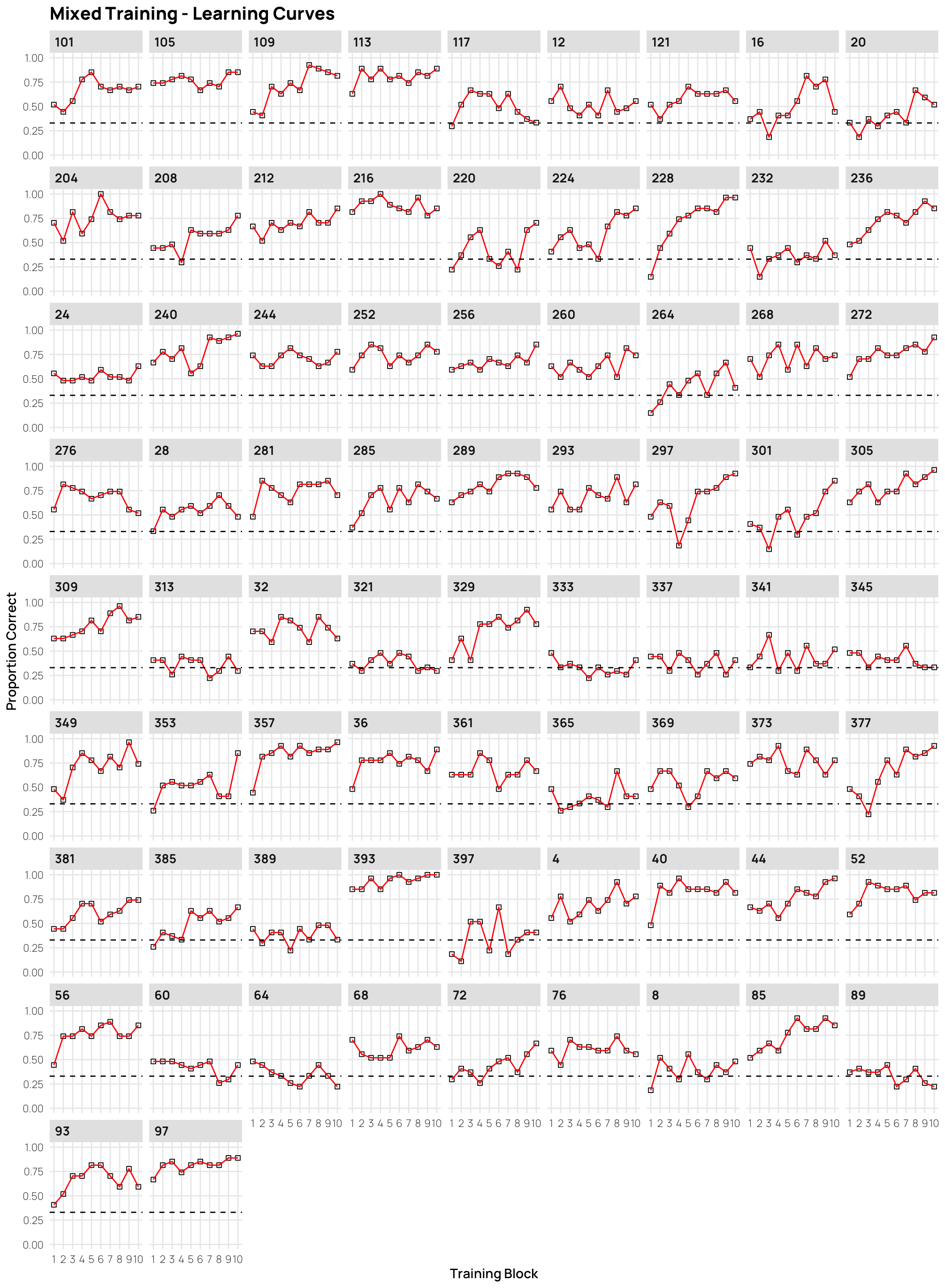
- facets sorted by final training accuracy
- click on plots to enlarge.
Display code
dCat |> filter(condit=="high", Phase==1) |>
ggplot(aes(x=Block, y=Corr)) +
stat_summary(shape=0,geom="point",fun="mean")+
stat_summary(geom="line",fun="mean",col="red")+
facet_wrap(~sbjCode)+ylim(c(0,1))+
geom_hline(yintercept = .33,linetype="dashed")+
ggtitle("High Training - Learning Curves")+
xlab("Training Block")+ylab("Proportion Correct")+scale_x_continuous(breaks=seq(1,10))Display code
dCat |> filter(condit=="low", Phase==1) |>
ggplot(aes(x=Block, y=Corr)) +
stat_summary(shape=0,geom="point",fun="mean")+
stat_summary(geom="line",fun="mean",col="red")+
facet_wrap(~sbjCode)+ylim(c(0,1))+
geom_hline(yintercept = .33,linetype="dashed")+
ggtitle("Low Training - Learning Curves")+
xlab("Training Block")+ylab("Proportion Correct")+scale_x_continuous(breaks=seq(1,10))Display code
dCat |> filter(condit=="medium", Phase==1) |>
ggplot(aes(x=Block, y=Corr)) +
stat_summary(shape=0,geom="point",fun="mean")+
stat_summary(geom="line",fun="mean",col="red")+
facet_wrap(~sbjCode)+ylim(c(0,1))+
geom_hline(yintercept = .33,linetype="dashed")+
ggtitle("Medium Training - Learning Curves")+
xlab("Training Block")+ylab("Proportion Correct")+scale_x_continuous(breaks=seq(1,10))Display code
dCat |> filter(condit=="mixed", Phase==1) |>
ggplot(aes(x=Block, y=Corr)) +
stat_summary(shape=0,geom="point",fun="mean")+
stat_summary(geom="line",fun="mean",col="red")+
facet_wrap(~sbjCode)+ylim(c(0,1))+
geom_hline(yintercept = .33,linetype="dashed")+
ggtitle("Mixed Training - Learning Curves")+
xlab("Training Block")+ylab("Proportion Correct")+scale_x_continuous(breaks=seq(1,10))Individual Testing
- facets sorted by final training accuracy
- click on plots to enlarge.
Display code
tx <- theme(axis.text.x=element_blank() )
dht <- dCat |> filter(condit=="high", Phase==2) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=Pattern_Token)) +
stat_summary(geom="bar",fun="mean")+
facet_wrap(~sbjCode, ncol=8)+
ylim(c(0,1))+
geom_hline(yintercept = .33,linetype="dashed", alpha=.5)+
ggtitle("High Distortion Training - Testing")+
xlab("Pattern Type")+ylab("Proportion Correct") +
theme(legend.position = "top") + tx
dlt <- dCat |> filter(condit=="low", Phase==2) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=Pattern_Token)) +
stat_summary(geom="bar",fun="mean")+
facet_wrap(~sbjCode, ncol=8)+
ylim(c(0,1))+
geom_hline(yintercept = .33,linetype="dashed", alpha=.5)+
ggtitle("Low Distortion Training - Testing")+
xlab("Pattern Type")+ylab("Proportion Correct")+
tx +theme(legend.position = "none")
dmt <- dCat |> filter(condit=="medium", Phase==2) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=Pattern_Token)) +
stat_summary(geom="bar",fun="mean")+
facet_wrap(~sbjCode, ncol=8)+
ylim(c(0,1))+
geom_hline(yintercept = .33,linetype="dashed", alpha=.5)+
ggtitle("Medium Distortion Training - Testing")+
xlab("Pattern Type")+ylab("Proportion Correct") +
theme(legend.position = "none")+
tx +theme(legend.position = "none")
dmxt <- dCat |> filter(condit=="mixed", Phase==2) |>
ggplot(aes(x=Pattern_Token, y=Corr, fill=Pattern_Token)) +
stat_summary(geom="bar",fun="mean")+
facet_wrap(~sbjCode, ncol=8)+
ylim(c(0,1))+
geom_hline(yintercept = .33,linetype="dashed", alpha=.5)+
ggtitle("Mixed Distortion Training - Testing")+
xlab("Pattern Type")+ylab("Proportion Correct")+
tx +theme(legend.position = "none")
(dht + dlt)/(dmt+dmxt)R packages used
Display code
# pkgs <- grateful::cite_packages(output = "table", pkgs="Session",out.dir = "assets", cite.tidyverse=TRUE)
# knitr::kable(pkgs)
#
# grateful::cite_packages(output = "paragraph",pkgs="Session",
# out.dir = "assets", cite.tidyverse=TRUE)
#
#
# pkgs <- grateful::cite_packages(output = "table",pkgs="Session",
# out.dir = "assets", cite.tidyverse=TRUE)
# knitr::kable(pkgs)
# #
# #
# pkgs <- cite_packages(cite.tidyverse = TRUE,
# output = "table",
# bib.file = "grateful-refs.bib",
# include.RStudio = TRUE,
# omit=c("colorout","viridis"),
# out.dir = getwd())
# formattable::formattable(pkgs,
# table.attr = 'class=\"table table-striped\" style="font-size: 14px; font-family: Lato; width: 80%"')
options(renv.config.dependencies.limit = Inf)
pkgs <- suppressWarnings(scan_packages(pkgs="Session",cite.tidyverse = TRUE))
knitr::kable(pkgs)| pkg | version |
|---|---|
| base | 4.4.0 |
| colorout | 1.3.0.2 |
| here | 1.0.1 |
| knitr | 1.47 |
| patchwork | 1.2.0 |
| tidyverse | 2.0.0 |
Display code
usedthese::used_here()| Package | Function |
|---|---|
| base | c[16], cut[2], library[1], list[8], max[3], mean[18], min[1], options[1], round[7], seq[15], source[2], suppressWarnings[1] |
| conflicted | conflict_prefer_all[1] |
| dplyr | arrange[1], filter[58], group_by[32], mutate[2], n_distinct[4], select[1], slice[6], summarise[8], summarize[16] |
| ggdist | stat_pointinterval[2] |
| ggplot2 | aes[58], element_blank[21], facet_wrap[25], geom_boxplot[2], geom_hline[14], geom_jitter[2], geom_smooth[4], ggplot[56], ggtitle[9], labs[47], position_dodge[66], position_jitter[2], position_jitterdodge[2], position_nudge[2], scale_x_continuous[6], scale_y_continuous[8], stat_summary[88], theme[23], theme_set[1], xlab[9], ylab[9], ylim[8] |
| grateful | scan_packages[1] |
| here | here[2] |
| knitr | kable[2] |
| pacman | p_load[1] |
| patchwork | plot_annotation[6], plot_layout[1] |
| stats | median[5] |
| stringr | str_wrap[6] |
| usedthese | used_here[1] |
Display code
#SystemInfo()