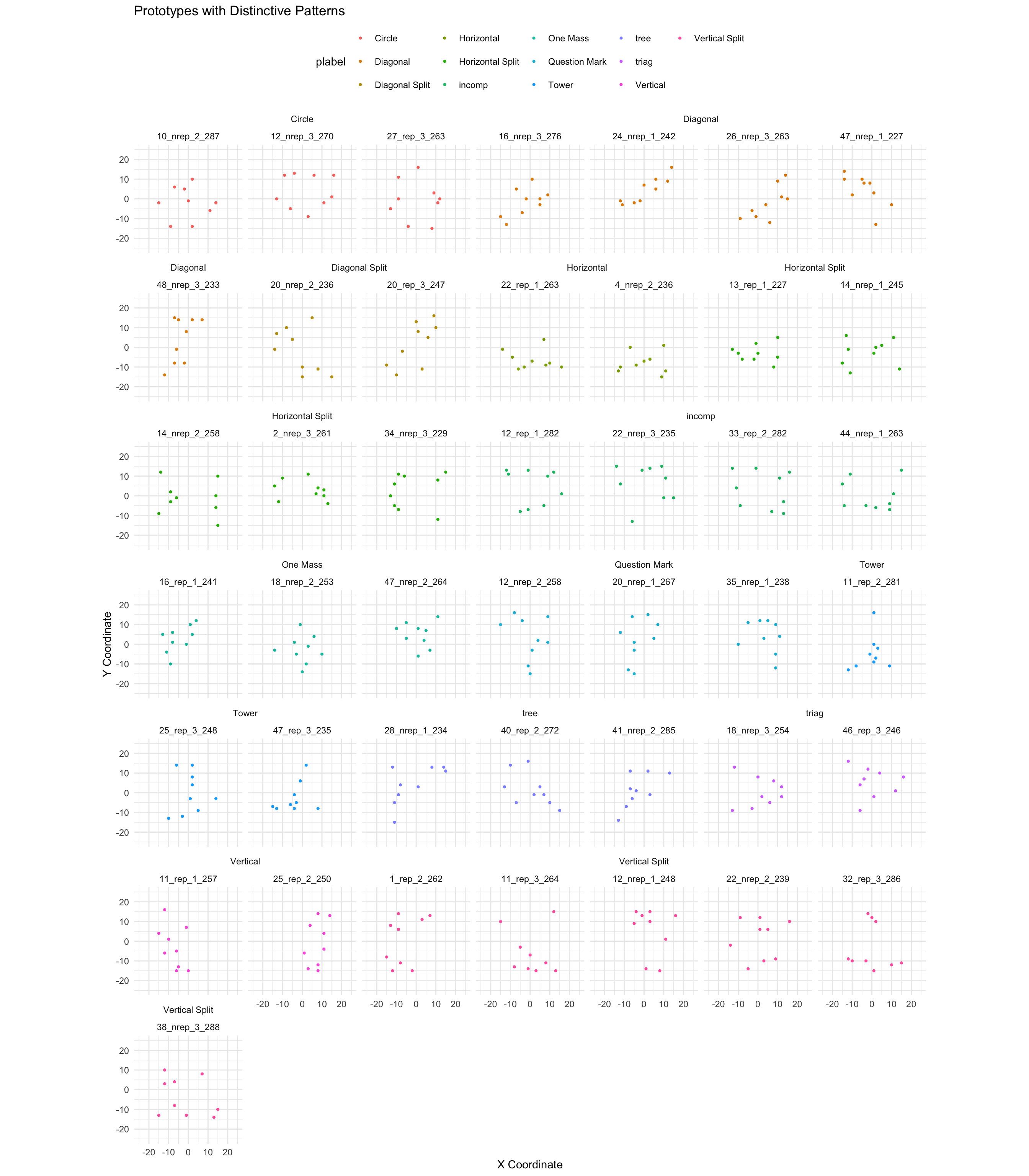
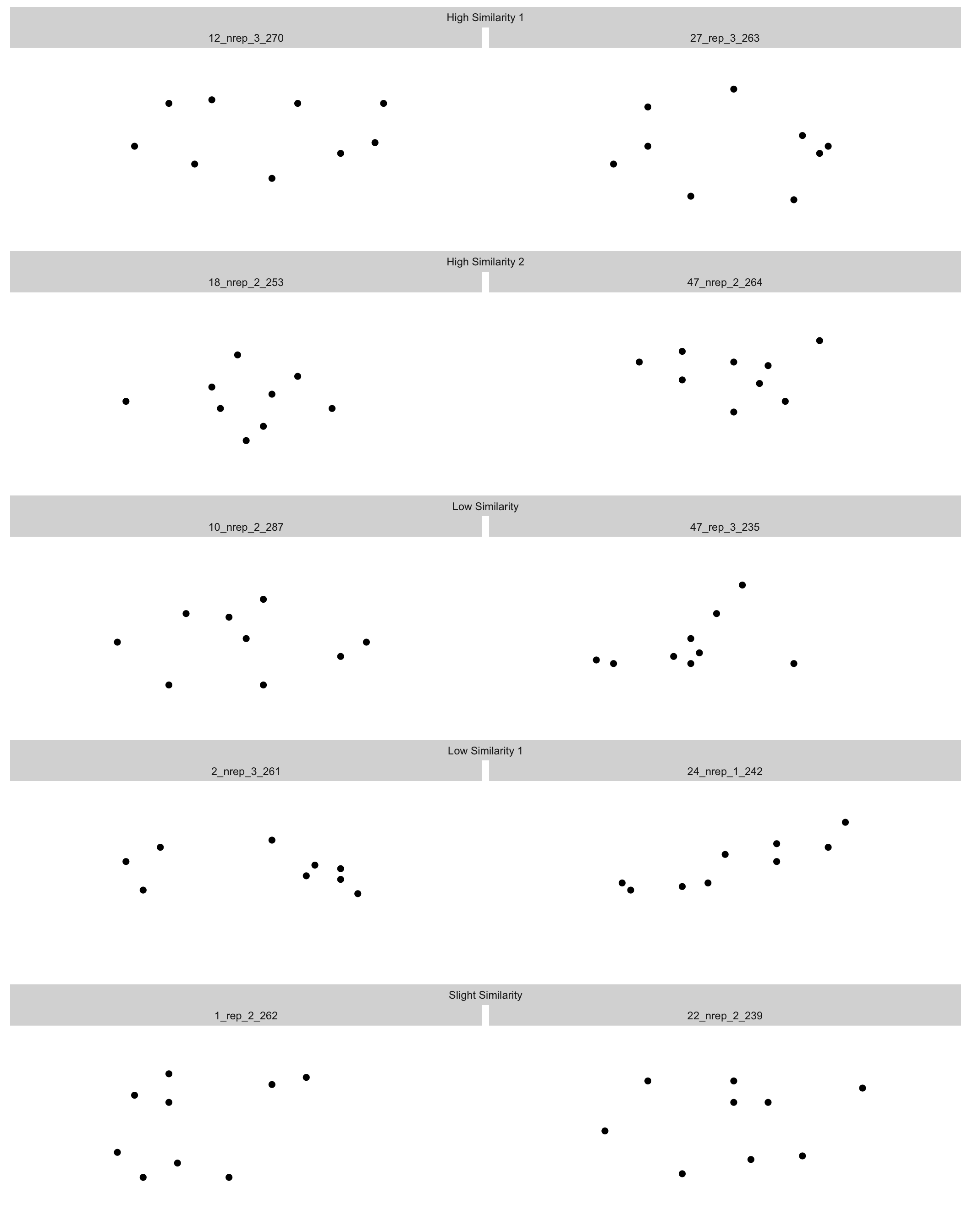
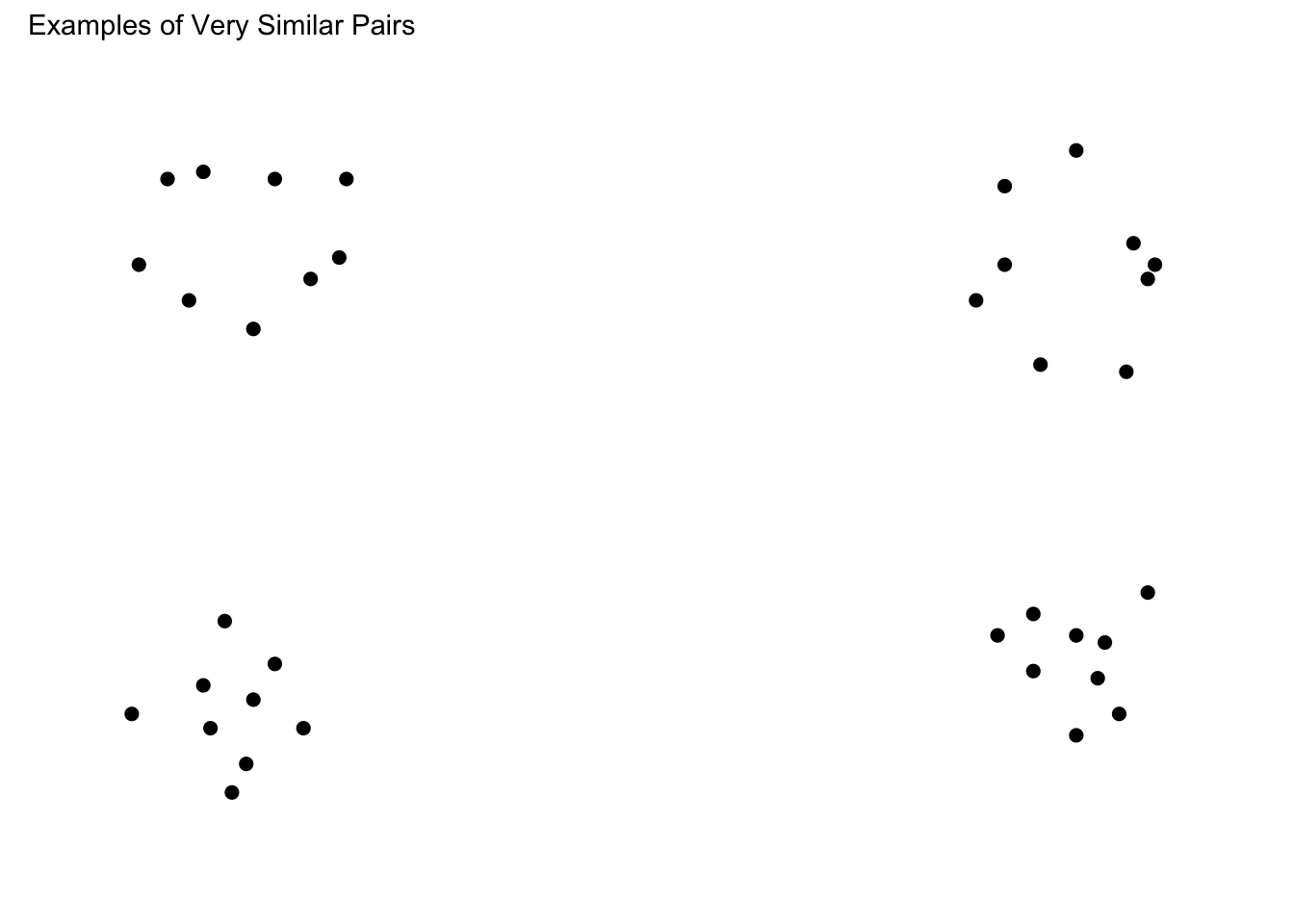
--- title: Dot Pattern Plots date: last-modified #page-layout: full format: html: grid: sidebar-width: 230px body-width: 1200px margin-width: 150px gutter-width: 1.0rem code-fold: true code-tools: true fig.width: 14 execute: warning: false eval: true --- ## Find Instruction Patterns ```{r} :: p_load (dplyr,purrr,tidyr,ggplot2, data.table,readr,here, patchwork, conflicted)conflict_prefer_all ("dplyr" , quiet = TRUE )# lmc22 <- readRDS(here("data","lmc22.rds")) # mc24 <- readRDS(here("data","mc24.rds")) <- read.csv (here ("Stimulii" ,"lmc22_prototypes.csv" ))<- read.csv (here ("Stimulii" ,"mc24_prototypes.csv" ))<- list (theme_minimal (),xlim (- 25 , 25 ),ylim (- 25 , 25 ),labs (x = "X Coordinate" , y = "Y Coordinate" ),coord_fixed (),guides (alpha = FALSE ))``` ### View large set of protypes from Hu & Nosofsky 2022 ```{r} #| fig-cap: Prototypes and their distortions #| fig-width: 11 #| fig-height: 13 <- lmc22_prototypes |> select (item_label, x1: y9) |> group_by (item_label) |> gather (key = "coordinate" , value = "value" , - item_label) %>% separate (coordinate, into = c ("axis" , "number" ), sep = 1 ) %>% spread (key = axis, value = value) %>% mutate (number = as.integer (number))|> filter (item_label %in% unique (proto_long$ item_label)[1 : 100 ]) |> ggplot (aes (x = x, y = y)) + geom_point (size= .75 ) + :: facet_nested_wrap (~ item_label) + + labs (title= "Prototypes from Category 1 - with distortions" )``` ### Prototypes with distinctive patterns ```{r} #| fig-cap: Prototypes with distinctive patterns #| fig-width: 14 #| fig-height: 16 <- c ("10_nrep_2_287" ,"12_nrep_3_270" , "27_rep_3_263" )<- c ("14_nrep_2_258" ,"13_rep_1_227" , "2_nrep_3_261" , "14_nrep_1_245" ,"34_nrep_3_229" )<- c ("1_rep_2_262" , "22_nrep_2_239" , "12_nrep_1_248" , "11_rep_3_264" ,"38_nrep_3_288" ,"32_rep_3_286" )<- c ("20_nrep_2_236" , "20_rep_3_247" )<- c ("11_rep_2_281" ,"25_rep_3_248" ,"47_rep_3_235" )<- c ("28_nrep_1_234" ,"41_nrep_2_285" ,"40_rep_2_272" )<- c ("12_nrep_2_258" ,"20_nrep_1_267" ,"35_nrep_1_238" )<- c ("24_nrep_1_242" ,"16_nrep_3_276" , "26_nrep_3_263" ,"48_nrep_3_233" ,"47_nrep_1_227" )<- c ("11_rep_1_257" , "25_rep_2_250" ,"48_nrep_3_233" )<- c ("22_rep_1_263" ,"4_nrep_2_236" )<- c ("13_rep_1_227" , "16_rep_1_241" ,"18_nrep_2_253" ,"47_nrep_2_264" )<- c ("18_nrep_3_254" ,"46_rep_3_246" )<- c ("22_nrep_3_235" ,"12_rep_1_282" ,"33_rep_2_282" , "44_nrep_1_263" )|> filter (item_label %in% c (circle, hsplit, vsplit, dsplit, tower, |> mutate (plabel = case_when (%in% circle ~ "Circle" ,%in% hsplit ~ "Horizontal Split" ,%in% vsplit ~ "Vertical Split" ,%in% dsplit ~ "Diagonal Split" ,%in% tower ~ "Tower" ,%in% triag ~ "triag" ,%in% tree ~ "tree" ,%in% triag ~ "Trianglish" ,%in% qmark ~ "Question Mark" ,%in% diag ~ "Diagonal" ,%in% vert ~ "Vertical" ,%in% horiz ~ "Horizontal" ,%in% incomp ~ "incomp" ,%in% onemass ~ "One Mass" )) |> ggplot (aes (x = x, y = y,col= plabel)) + geom_point (size= .75 ) + :: facet_nested_wrap (~ plabel+ item_label) + + labs (title= "Prototypes with Distinctive Patterns" ) + theme (legend.position = "top" )``` ### Organize subset into pairs ```{r} #| fig-cap: Prototype pairs with different similarity levels #| fig-width: 11 #| fig-height: 14 <- circle[2 : 3 ]<- onemass[3 : 4 ]<- vsplit[1 : 2 ]<- c (diag[1 ],hsplit[3 ])<- c (circle[1 ],tower[3 ])|> filter (item_label %in% c (hsim1, hsim2, msim1, lsim1, lsim2)) |> mutate (plabel = case_when (%in% hsim1 ~ "High Similarity 1" ,%in% hsim2 ~ "High Similarity 2" ,%in% msim1 ~ "Slight Similarity" ,%in% lsim1 ~ "Low Similarity 1" ,%in% lsim2 ~ "Low Similarity" )) |> ggplot (aes (x = x, y = y)) + geom_point (size= 2 ) + :: facet_nested_wrap (~ plabel+ item_label,ncol= 2 ) + theme (panel.background = element_blank (),axis.text = element_blank (),axis.title = element_blank (),axis.ticks = element_blank ()) + xlim (- 25 , 25 ) + ylim (- 25 ,25 )``` ### Finalize and save patterns ```{r} <- list ( theme (panel.background = element_blank (),axis.text = element_blank (),axis.title = element_blank (),axis.ticks = element_blank (),panel.grid = element_blank (),# no facet labels strip.background = element_blank (),strip.text.x = element_blank (),panel.spacing = unit (12 , "lines" )), xlim (- 25 , 25 ), ylim (- 25 ,25 ))<- proto_long |> filter (item_label %in% hsim1) |> ggplot (aes (x = x, y = y)) + geom_point (size= 2 ) + facet_wrap (~ item_label) + <- proto_long |> filter (item_label %in% hsim2) |> ggplot (aes (x = x, y = y)) + geom_point (size= 2 ) + facet_wrap (~ item_label) + <- proto_long |> filter (item_label %in% msim1) |> ggplot (aes (x = x, y = y)) + geom_point (size= 2 ) + facet_wrap (~ item_label) + <- proto_long |> filter (item_label %in% lsim1) |> ggplot (aes (x = x, y = y)) + geom_point (size= 2 ) + facet_wrap (~ item_label) + <- proto_long |> filter (item_label %in% lsim2) |> ggplot (aes (x = x, y = y)) + geom_point (size= 2 ) + facet_wrap (~ item_label) + # phsim1 # phsim2 # pmsim1 # plsim1 # plsim2 <- (phsim1/ phsim2) <- (plsim1/ plsim2)+ plot_annotation (subtitle = 'Examples of Very Similar Pairs' )+ plot_annotation (subtitle = 'Examples of Not Similar Pairs' )# save_plots # ggsave(here("Task/assets/high_sim.png"),very_similar) # ggsave(here("Task/assets/low_sim.png"),not_similar) ``` # save examples separately ```{r} # ggsave(here("Task/assets/high_sim1.png"),phsim1) # ggsave(here("Task/assets/high_sim2.png"),phsim2) # # ggsave(here("Task/assets/low_sim1.png"),plsim1) # ggsave(here("Task/assets/low_sim2.png"),plsim2) ```